1PER
 
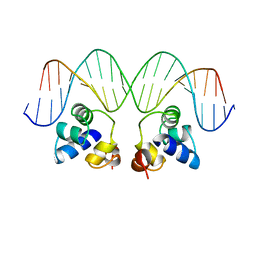 | |
1KQR
 
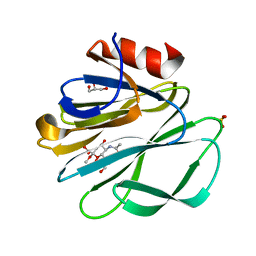 | | Crystal Structure of the Rhesus Rotavirus VP4 Sialic Acid Binding Domain in Complex with 2-O-methyl-alpha-D-N-acetyl neuraminic acid | | Descriptor: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Dormitzer, P.R, Sun, Z.-Y.J, Wagner, G, Harrison, S.C. | | Deposit date: | 2002-01-07 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Rhesus Rotavirus VP4 Sialic Acid Binding Domain has a Galectin Fold with a
Novel Carbohydrate Binding Site
Embo J., 21, 2002
|
|
9DOF
 
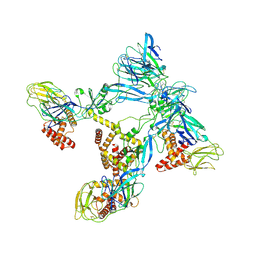 | | Octahedral small virus-like particles of dengue virus type 2 (local reconstruction) | | Descriptor: | Protein prM, glycoprotein E | | Authors: | Johnson, A, Dodes Traian, M, Walsh, R.M, Jenni, S, Harrison, S.C. | | Deposit date: | 2024-09-19 | | Release date: | 2024-12-11 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Octahedral small virus-like particles of dengue virus type 2.
J.Virol., 99, 2025
|
|
9DOG
 
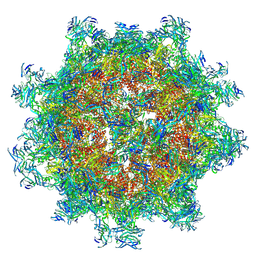 | | Octahedral small virus-like particles of dengue virus type 2 (octahedral reconstruction) | | Descriptor: | Protein prM, glycoprotein E | | Authors: | Johnson, A, Dodes Traian, M, Walsh, R.M, Jenni, S, Harrison, S.C. | | Deposit date: | 2024-09-19 | | Release date: | 2024-12-11 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Octahedral small virus-like particles of dengue virus type 2.
J.Virol., 99, 2025
|
|
5A22
 
 | | Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy | | Descriptor: | VESICULAR STOMATITIS VIRUS L POLYMERASE, ZINC ION | | Authors: | Liang, B, Li, Z, Jenni, S, Rameh, A.A, Morin, B.M, Grant, T, Grigorieff, N, Harrison, S.C, Whelan, S.P.J. | | Deposit date: | 2015-05-06 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the L Protein of Vesicular Stomatitis Virus from Electron Cryomicroscopy.
Cell(Cambridge,Mass.), 162, 2015
|
|
2IGP
 
 | | Crystal Structure of Hec1 CH domain | | Descriptor: | BETA-MERCAPTOETHANOL, Retinoblastoma-associated protein HEC | | Authors: | Wei, R.R, Harrison, S.C. | | Deposit date: | 2006-09-22 | | Release date: | 2007-01-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment.
Nat.Struct.Mol.Biol., 14, 2007
|
|
3FMG
 
 | | Structure of rotavirus outer capsid protein VP7 trimer in complex with a neutralizing Fab | | Descriptor: | CALCIUM ION, Fab of neutralizing antibody 4F8, heavy chain, ... | | Authors: | Aoki, S.T, Settembre, E.C, Trask, S.D, Greenberg, H.B, Harrison, S.C, Dormitzer, P.R. | | Deposit date: | 2008-12-22 | | Release date: | 2009-06-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of rotavirus outer-layer protein VP7 bound with a neutralizing Fab.
Science, 324, 2009
|
|
4QHL
 
 | | I3.2 (unbound) from CH103 Lineage | | Descriptor: | I3 heavy chain, UCA light chain | | Authors: | Fera, D, Harrison, S.C. | | Deposit date: | 2014-05-28 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.153 Å) | | Cite: | Affinity maturation in an HIV broadly neutralizing B-cell lineage through reorientation of variable domains.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QHN
 
 | | I2 (unbound) from CH103 Lineage | | Descriptor: | I2 heavy chain, I2 light chain | | Authors: | Fera, D, Harrison, S.C. | | Deposit date: | 2014-05-28 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Affinity maturation in an HIV broadly neutralizing B-cell lineage through reorientation of variable domains.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4QHM
 
 | | I3.1 (unbound) from CH103 Lineage | | Descriptor: | I2 light chain, I3 heavy chain | | Authors: | Fera, D, Harrison, S.C. | | Deposit date: | 2014-05-28 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Affinity maturation in an HIV broadly neutralizing B-cell lineage through reorientation of variable domains.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
9C1I
 
 | |
9C1G
 
 | | Rhesus rotavirus (consensus structure at 2.36 Angstrom resolution) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Jenni, S, Herrmann, T, De Sautu, M, Harrison, S.C. | | Deposit date: | 2024-05-29 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Rotavirus structure
To Be Published
|
|
9C1H
 
 | | Rhesus rotavirus (upright structure at 2.88 Angstrom resolution) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Jenni, S, Herrmann, T, De Sautu, M, Harrison, S.C. | | Deposit date: | 2024-05-29 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Rotavirus structure
To Be Published
|
|
9C1L
 
 | |
9C1K
 
 | | Rhesus rotavirus (empty structure at 2.68 Angstrom resolution) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Jenni, S, Herrmann, T, De Sautu, M, Harrison, S.C. | | Deposit date: | 2024-05-29 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Rotavirus structure
To Be Published
|
|
9C1J
 
 | | Rhesus rotavirus (reversed structure at 2.72 Angstrom resolution) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Jenni, S, Herrmann, T, De Sautu, M, Harrison, S.C. | | Deposit date: | 2024-05-29 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Rotavirus structure
To Be Published
|
|
4QHK
 
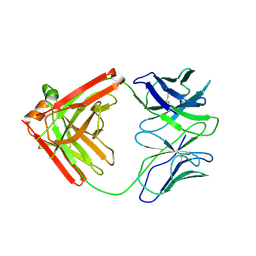 | | UCA (unbound) from CH103 Lineage | | Descriptor: | UCA heavy chain, UCA light chain | | Authors: | Fera, D, Harrison, S.C. | | Deposit date: | 2014-05-28 | | Release date: | 2014-06-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.487 Å) | | Cite: | Affinity maturation in an HIV broadly neutralizing B-cell lineage through reorientation of variable domains.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1R69
 
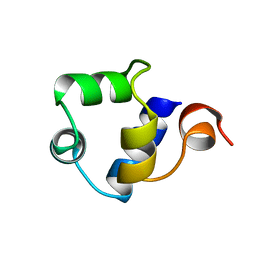 | | STRUCTURE OF THE AMINO-TERMINAL DOMAIN OF PHAGE 434 REPRESSOR AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | REPRESSOR PROTEIN CI | | Authors: | Mondragon, A, Subbiah, S, Alamo, S.C, Drottar, M, Harrison, S.C. | | Deposit date: | 1988-12-08 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the amino-terminal domain of phage 434 repressor at 2.0 A resolution.
J.Mol.Biol., 205, 1989
|
|
2CRO
 
 | |
2FTX
 
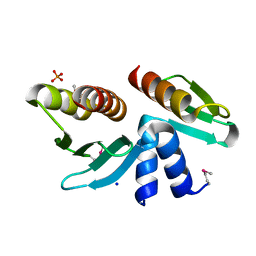 | | Crystal structure of the yeast kinetochore Spc24/Spc25 globular domain | | Descriptor: | Hypothetical 24.6 kDa protein in ILV2-ADE17 intergenic region, Hypothetical 25.2 kDa protein in AFG3-SEB2 intergenic region, PHOSPHATE ION, ... | | Authors: | Wei, R.R, Harrison, S.C. | | Deposit date: | 2006-01-25 | | Release date: | 2006-06-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Atomic Structure of the kinetochore Spc24p/Spc25p globular domain reveals a novel fold
To be Published
|
|
2YFW
 
 | |
1PZU
 
 | | An asymmetric NFAT1-RHR homodimer on a pseudo-palindromic, Kappa-B site | | Descriptor: | 5'-D(*AP*AP*TP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*C)-3', 5'-D(*TP*TP*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*A)-3', Nuclear factor of activated T-cells, ... | | Authors: | Jin, L, Sliz, P, Chen, L, Macian, F, Rao, A, Hogan, P.G, Harrison, S.C. | | Deposit date: | 2003-07-14 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An asymmetric NFAT1 dimer on a pseudo-palindromic KB-like DNA site
Nat.Struct.Biol., 10, 2003
|
|
1PYI
 
 | |
1SVA
 
 | | SIMIAN VIRUS 40 | | Descriptor: | SIMIAN VIRUS 40 | | Authors: | Stehle, T, Gamblin, S.J, Harrison, S.C. | | Deposit date: | 1995-11-27 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of simian virus 40 refined at 3.1 A resolution.
Structure, 4, 1996
|
|
2OR1
 
 | | RECOGNITION OF A DNA OPERATOR BY THE REPRESSOR OF PHAGE 434. A VIEW AT HIGH RESOLUTION | | Descriptor: | 434 REPRESSOR, DNA (5'-D(*AP*AP*GP*TP*AP*CP*AP*AP*AP*CP*TP*TP*TP*CP*TP*TP*G P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*CP*AP*AP*GP*AP*AP*AP*GP*TP*TP*TP*GP*T P*AP*CP*T)-3') | | Authors: | Aggarwal, A.K, Rodgers, D.W, Drottar, M, Ptashne, M, Harrison, S.C. | | Deposit date: | 1989-09-05 | | Release date: | 1989-09-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Recognition of a DNA operator by the repressor of phage 434: a view at high resolution.
Science, 242, 1988
|
|
