2JGP
 
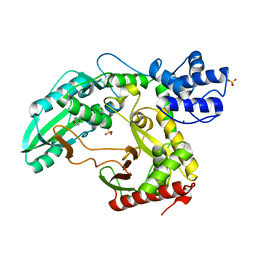 | | Structure of the TycC5-6 PCP-C bidomain of the tyrocidine synthetase TycC | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, SODIUM ION, SULFATE ION, ... | | Authors: | Samel, S.A, Schoenafinger, G, Knappe, T.A, Marahiel, M.A, Essen, L.-O. | | Deposit date: | 2007-02-13 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Insights Into a Peptide Bond-Forming Bidomain from a Nonribosomal Peptide Synthetase.
Structure, 15, 2007
|
|
1T3E
 
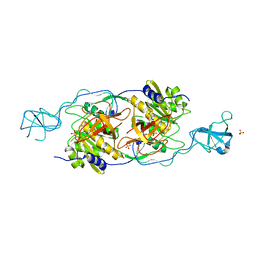 | | Structural basis of dynamic glycine receptor clustering | | Descriptor: | 49-mer fragment of Glycine receptor beta chain, Gephyrin, SULFATE ION | | Authors: | Sola, M, Bavro, V.N, Timmins, J, Franz, T, Ricard-Blum, S, Schoehn, G, Ruigrok, R.W.H, Paarmann, I, Saiyed, T, O'Sullivan, G.A. | | Deposit date: | 2004-04-26 | | Release date: | 2004-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural basis of dynamic glycine receptor clustering by gephyrin
Embo J., 23, 2004
|
|
2W1K
 
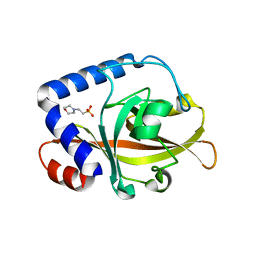 | | Crystal Structure of Sortase C-3 (SRTC-3) from Streptococcus pneumoniae | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, PUTATIVE SORTASE | | Authors: | Manzano, C, Contreras-Martel, C, El Mortaji, L, Izore, T, Fenel, D, Vernet, T, Schoehn, G, Di Guilmi, A.M, Dessen, A. | | Deposit date: | 2008-10-17 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Sortase-Mediated Pilus Fiber Biogenesis in Streptococcus Pneumoniae.
Structure, 16, 2008
|
|
1E97
 
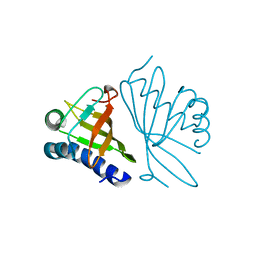 | |
1GS3
 
 | |
6K68
 
 | | Application of anti-helix antibodies in protein structure determination (8420-3MNZ) | | Descriptor: | 3MNZ Variable heavy chain, 3MNZ Variable light chain, Protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-01 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K69
 
 | | Application of anti-helix antibodies in protein structure determination (9213-3LRH) | | Descriptor: | 3LRH intrabody, Engineered T4 lysozyme | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-01 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8CHO
 
 | |
1DMQ
 
 | | CRYSTAL STRUCTURE OF MUTANT ENZYME Y32F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1DMN
 
 | | CRYSTAL STRUCTURE OF MUTANT ENZYME Y32F/Y57F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1DMM
 
 | | CRYSTAL STRUCTURES OF MUTANT ENZYMES Y57F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1W02
 
 | |
1W00
 
 | | Crystal structure of mutant enzyme D103L of Ketosteroid Isomerase from Pseudomonas putida biotype B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 2004-05-30 | | Release date: | 2005-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Double-Mutant Cycle Analysis of a Hydrogen Bond Network in Ketosteroid Isomerase from Pseudomonas Putida Biotype B
Biochem.J., 382, 2004
|
|
1W01
 
 | |
6K65
 
 | | Application of anti-helix antibodies in protein structure determination (9014-1P4B) | | Descriptor: | 1P4B variable heavy chain, 1P4B variable light chain, Immunoglobulin G-binding protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-01 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K6B
 
 | | Application of anti-helix antibodies in protein structure determination (8496-3LRH) | | Descriptor: | 3LRH intrabody, Protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-02 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K3M
 
 | | Application of anti-helix antibodies in protein structure determination (8189-3LRH) | | Descriptor: | 3LRH intrabody, SpA IgG-binding domain protein,Protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-05-20 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K67
 
 | | Application of anti-helix antibodies in protein structure determination (9011-3LRH) | | Descriptor: | 3LRH introbody, CALCIUM ION, Engineered calmodulin | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-01 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K64
 
 | | Application of anti-helix antibodies in protein structure determination (8188-3LRH) | | Descriptor: | 3LRH intrabody, Protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-01 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K6A
 
 | | Application of anti-helix antibodies in protein structure determination (8188cys-3LRHcys) | | Descriptor: | 3LRH intrabody, Engineered Protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-02 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1QJG
 
 | |
5AFX
 
 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-1238 | | Descriptor: | FARNESYL PYROPHOSPHATE SYNTHASE, MAGNESIUM ION, [1-hydroxy-2-(1-nonyl-1H-3lambda~5~-imidazol-3-yl)ethane-1,1-diyl]bis(phosphonic acid) | | Authors: | Yang, G, Oldfield, E, No, J.H. | | Deposit date: | 2015-01-27 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Inhibition of Trypanosoma Brucei Cell Growth by Lipophilic Bisphosphonates: An in Vitro and in Vivo Investigation.
Antimicrob.Agents Chemother., 59, 2015
|
|
5AEL
 
 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-597 | | Descriptor: | FARNESYL PYROPHOSPHATE SYNTHASE, MAGNESIUM ION, {2-[3-(hex-1-yn-1-yl)pyridinium-1-yl]ethane-1,1-diyl}bis(phosphonate) | | Authors: | Yang, G, Oldfield, E, No, J.H. | | Deposit date: | 2014-12-26 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Inhibition of Trypanosoma Brucei Cell Growth by Lipophilic Bisphosphonates: An in Vitro and in Vivo Investigation.
Antimicrob.Agents Chemother., 59, 2015
|
|
5AHU
 
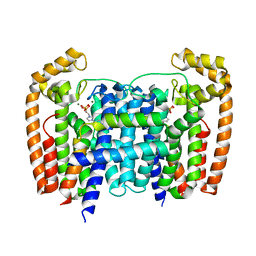 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-1326 | | Descriptor: | FARNESYL PYROPHOSPHATE SYNTHASE, PUTATIVE, MAGNESIUM ION, ... | | Authors: | Yang, G, Oldfield, E, No, J.H. | | Deposit date: | 2015-02-09 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Inhibition of Trypanosoma Brucei Cell Growth by Lipophilic Bisphosphonates: An in Vitro and in Vivo Investigation.
Antimicrob.Agents Chemother., 59, 2015
|
|
3NBX
 
 | |
