3V6A
 
 | |
5J1M
 
 | | Crystal structure of Csd1-Csd2 dimer II | | Descriptor: | ToxR-activated gene (TagE), ZINC ION | | Authors: | An, D.R, Suh, S.W. | | Deposit date: | 2016-03-29 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of the Heterodimer Formation between Cell Shape-Determining Proteins Csd1 and Csd2 from Helicobacter pylori
Plos One, 11, 2016
|
|
5J1K
 
 | | Crystal structure of Csd2-Csd2 dimer | | Descriptor: | GLYCEROL, ToxR-activated gene (TagE) | | Authors: | An, D.R, Suh, S.W. | | Deposit date: | 2016-03-29 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Basis of the Heterodimer Formation between Cell Shape-Determining Proteins Csd1 and Csd2 from Helicobacter pylori
Plos One, 11, 2016
|
|
5J1L
 
 | | Crystal structure of Csd1-Csd2 dimer I | | Descriptor: | ToxR-activated gene (TagE), ZINC ION | | Authors: | An, D.R, Suh, S.W. | | Deposit date: | 2016-03-29 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Basis of the Heterodimer Formation between Cell Shape-Determining Proteins Csd1 and Csd2 from Helicobacter pylori
Plos One, 11, 2016
|
|
7CC7
 
 | |
7EC3
 
 | | Crystal structure of SdgB (complexed with UDP, GlcNAc, and Glycosylated peptide) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-35)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-65)]5,6-DIHYDRO-BENZO[H]CINNOLIN-3-YLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosyl transferase, ... | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7EC6
 
 | | Crystal structure of SdgB (complexed with peptides) | | Descriptor: | ASP-SER-ASP, Glycosyl transferase, group 1 family protein | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7EC7
 
 | | Crystal structure of SdgB (complexed with phosphate ions) | | Descriptor: | Glycosyl transferase, group 1 family protein, PHOSPHATE ION | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7EC1
 
 | | Crystal structure of SdgB (ligand-free form) | | Descriptor: | GLYCEROL, Glycosyl transferase, group 1 family protein, ... | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-03-11 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7EWJ
 
 | | Toxin-antitoxin complex from Staphylococcus aureus | | Descriptor: | Endoribonuclease MazF, GLYCEROL, PemI inhibitor, ... | | Authors: | Kim, D.H, Kang, S.M, Lee, S.J, Lee, B.J. | | Deposit date: | 2021-05-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of PemI in the Staphylococcus aureus PemIK toxin-antitoxin complex: PemI controls PemK by acting as a PemK loop mimic.
Nucleic Acids Res., 50, 2022
|
|
7EWI
 
 | | Toxin protein from Staphylococcus aureus | | Descriptor: | Endoribonuclease MazF, GLYCEROL, PHOSPHATE ION | | Authors: | Kim, D.H, Kang, S.M, Lee, S.J, Lee, B.J. | | Deposit date: | 2021-05-25 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Role of PemI in the Staphylococcus aureus PemIK toxin-antitoxin complex: PemI controls PemK by acting as a PemK loop mimic.
Nucleic Acids Res., 50, 2022
|
|
7ERR
 
 | |
6JHW
 
 | | Structure of anti-CRISPR AcrIIC3 and NmeCas9 HNH | | Descriptor: | AcrIIC3, CRISPR-associated endonuclease Cas9 | | Authors: | Suh, J.Y, Lee, B.J, Lee, S.J, Kim, Y. | | Deposit date: | 2019-02-19 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Anti-CRISPR AcrIIC3 discriminates between Cas9 orthologs via targeting the variable surface of the HNH nuclease domain.
Febs J., 286, 2019
|
|
6JHV
 
 | | Structure of anti-CRISPR AcrIIC3 | | Descriptor: | AcrIIC3 | | Authors: | Suh, J.Y, Lee, B.J, Lee, S.J, Kim, Y. | | Deposit date: | 2019-02-19 | | Release date: | 2019-08-28 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.321 Å) | | Cite: | Anti-CRISPR AcrIIC3 discriminates between Cas9 orthologs via targeting the variable surface of the HNH nuclease domain.
Febs J., 286, 2019
|
|
6IFC
 
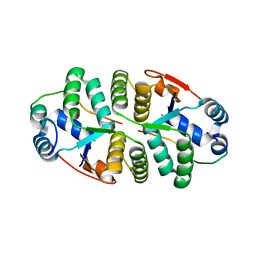 | | Crystal structure of VapBC from Salmonella typhimurium | | Descriptor: | Antitoxin VapB, CALCIUM ION, tRNA(fMet)-specific endonuclease VapC | | Authors: | Park, D.W, Lee, B.J. | | Deposit date: | 2018-09-19 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of proteolyzed VapBC and DNA-bound VapBC from Salmonella enterica Typhimurium LT2 and VapC as a putative Ca2+-dependent ribonuclease.
Faseb J., 34, 2020
|
|
6IFM
 
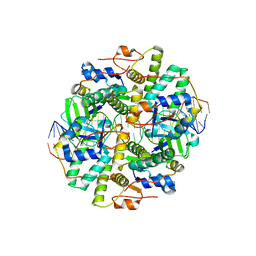 | | Crystal structure of DNA bound VapBC from Salmonella typhimurium | | Descriptor: | Antitoxin VapB, DNA backward (27-MER), DNA forward (27-MER), ... | | Authors: | Park, D.W, Lee, B.J. | | Deposit date: | 2018-09-20 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Crystal structure of proteolyzed VapBC and DNA-bound VapBC from Salmonella enterica Typhimurium LT2 and VapC as a putative Ca2+-dependent ribonuclease.
Faseb J., 34, 2020
|
|
5YRZ
 
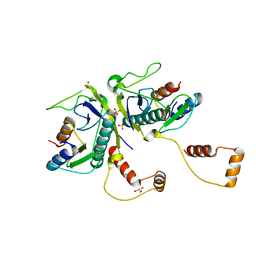 | | Toxin-Antitoxin complex from Streptococcus pneumoniae | | Descriptor: | GLYCEROL, HicA, HicB, ... | | Authors: | Kang, S.M, Kim, D.H. | | Deposit date: | 2017-11-11 | | Release date: | 2018-05-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Functional insights into the Streptococcus pneumoniae HicBA toxin-antitoxin system based on a structural study.
Nucleic Acids Res., 46, 2018
|
|
4NRN
 
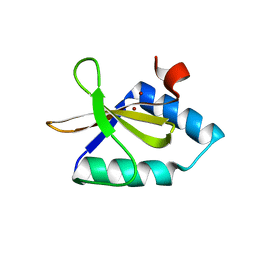 | | Crystal structure of metal-bound toxin from Helicobacter pylori | | Descriptor: | ZINC ION, metal-bound toxin | | Authors: | Lee, B.J, Im, H, Pathak, C, Jang, S.B. | | Deposit date: | 2013-11-27 | | Release date: | 2014-10-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal structure of toxin HP0892 from Helicobacter pylori with two Zn(II) at 1.8 angstrom resolution
Protein Sci., 23, 2014
|
|
4NJQ
 
 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CARBONATE ION, COBALT (II) ION, ... | | Authors: | Nguyen, D.D, Pandian, R, Kim, D.Y, Ha, S.C, Yun, K.H, Kim, K.S, Kim, J.H, Kim, K.K. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
7VFN
 
 | | Crystal structure of SdgB (SD peptide-binding form) | | Descriptor: | ASP-SER-ASP, Glycosyl transferase, group 1 family protein | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7VFM
 
 | | Crystal structure of SdgB (UDP and SD peptide-binding form) | | Descriptor: | Glycosyl transferase, group 1 family protein, SER-ASP-SER-ASP, ... | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7VFL
 
 | | Crystal structure of SdgB (UDP, NAG, and O-glycosylated SD peptide-binding form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosyl transferase, group 1 family protein, ... | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7VFO
 
 | | Crystal structure of SdgB (Phosphate-binding form) | | Descriptor: | Glycosyl transferase, group 1 family protein, PHOSPHATE ION | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7VFK
 
 | | Crystal structure of SdgB (ligand-free form) | | Descriptor: | GLYCEROL, Glycosyl transferase, group 1 family protein, ... | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7XDP
 
 | |
