3KUE
 
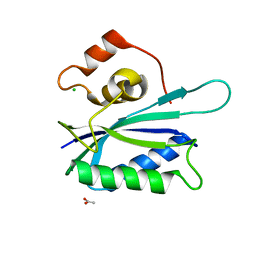 | | Crystal structure of E. coli HPPK(E77A) | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, CHLORIDE ION | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-11-27 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.538 Å) | | Cite: | Roles of residues E77 and H115 in E. coli HPPK
To be Published
|
|
3KUG
 
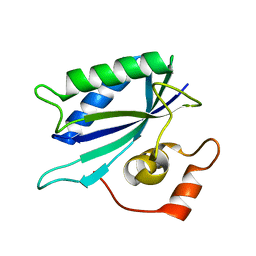 | |
3KUH
 
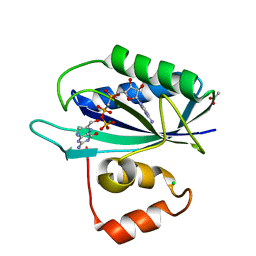 | | Crystal structure of E. coli HPPK(H115A) in complex with AMPCPP and HP | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-11-27 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Roles of residues E77 and H115 in E. coli HPPK
To be Published
|
|
7P88
 
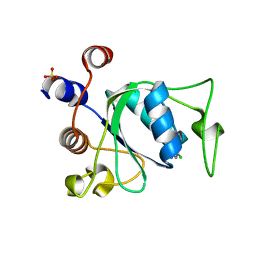 | | Crystal structure of YTHDC1 with compound YLI_DC1_002 | | Descriptor: | 2-chloranyl-~{N}-methyl-9~{H}-purin-6-amine, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Li, Y, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
7P8B
 
 | | Crystal structure of YTHDC1 with compound YLI_DC1_006 | | Descriptor: | 9-cyclopropyl-~{N}-methyl-purin-6-amine, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Li, Y, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
7P8A
 
 | | Crystal structure of YTHDC1 with compound YLI_DC1_003 | | Descriptor: | SULFATE ION, YTH domain-containing protein 1, ~{N},9-dimethylpurin-6-amine | | Authors: | Bedi, R.K, Li, Y, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
7P87
 
 | | Crystal structure of YTHDC1 with compound YLI_DC1_001 | | Descriptor: | N-methyl-4,5,6,7-tetrahydro-1H-indazole-3-carboxamide, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Li, Y, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
7P8F
 
 | | Crystal structure of YTHDC1 with compound YLI_DC1_008 | | Descriptor: | SULFATE ION, YTH domain-containing protein 1, ~{N},3-dimethyl-1~{H}-pyrazolo[4,3-d]pyrimidin-7-amine | | Authors: | Bedi, R.K, Li, Y, Caflisch, A. | | Deposit date: | 2021-07-21 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
7X8B
 
 | |
7X8G
 
 | |
7X8F
 
 | |
6L5Z
 
 | |
8ZPT
 
 | | Cryo-EM structure of prolactin-releasing peptide recognition with Gq | | Descriptor: | Guanine nucleotide-binding protein G(324) subunit alpha-1,, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, L, Li, Y, Yuan, Q, Xu, H.E. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Molecular mechanism of prolactin-releasing peptide recognition and signaling via its G protein-coupled receptor.
Cell Discov, 10, 2024
|
|
8ZPS
 
 | | Cryo-EM structure of prolactin-releasing peptide recognition with Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha-1, ... | | Authors: | Zhao, L, Li, Y, Yuan, Q, Xu, H.E. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Molecular mechanism of prolactin-releasing peptide recognition and signaling via its G protein-coupled receptor.
Cell Discov, 10, 2024
|
|
4TMP
 
 | | Crystal structure of AF9 YEATS bound to H3K9ac peptide | | Descriptor: | 1,2-ETHANEDIOL, ALA-ARG-THR-LYS-GLN-THR-ALA-ARG-ALY-SER-THR, Protein AF-9 | | Authors: | Li, H, Li, Y, Wang, H, Ren, Y. | | Deposit date: | 2014-06-02 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | AF9 YEATS Domain Links Histone Acetylation to DOT1L-Mediated H3K79 Methylation.
Cell, 159, 2014
|
|
6DZP
 
 | | Cryo-EM Structure of Mycobacterium smegmatis C(minus) 50S ribosomal subunit | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Sharma, M.R, Li, Y, Korripella, R, Yang, Y, Kaushal, P.S, Lin, Q, Wade, J.T, Gray, A.G, Derbyshire, K.M, Agrawal, R.K, Ojha, A. | | Deposit date: | 2018-07-05 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Zinc depletion induces ribosome hibernation in mycobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DZK
 
 | | Cryo-EM Structure of Mycobacterium smegmatis C(minus) 30S ribosomal subunit with MPY | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Sharma, M.R, Li, Y, Korripella, R, Yang, Y, Kaushal, P.S, Lin, Q, Wade, J.T, Gray, A.G, Derbyshire, K.M, Agrawal, R.K, Ojha, A. | | Deposit date: | 2018-07-05 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Zinc depletion induces ribosome hibernation in mycobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DZI
 
 | | Cryo-EM Structure of Mycobacterium smegmatis 70S C(minus) ribosome 70S-MPY complex | | Descriptor: | 16S rRNA, 23 S rRNA (3119-MER), 30S ribosomal protein S10, ... | | Authors: | Sharma, M.R, Li, Y, Korripella, R, Yang, Y, Kaushal, P.S, Lin, Q, Wade, J.T, Gray, A.G, Derbyshire, K.M, Agrawal, R.K, Ojha, A. | | Deposit date: | 2018-07-05 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Zinc depletion induces ribosome hibernation in mycobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5J9S
 
 | | ENL YEATS in complex with histone H3 acetylation at K27 | | Descriptor: | H3K27ac peptide from Histone H3.1, Protein ENL, SULFATE ION | | Authors: | Li, Y, Li, H. | | Deposit date: | 2016-04-11 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | ENL links histone acetylation to oncogenic gene expression in acute myeloid leukaemia
Nature, 543, 2017
|
|
4ZRK
 
 | | Merlin-FERM and Lats1 complex | | Descriptor: | Merlin, Serine/threonine-protein kinase LATS1 | | Authors: | Lin, Z, Li, Y, Wei, Z, Zhang, M. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.316 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
4N4G
 
 | |
4N4I
 
 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.3K36me3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.3, ... | | Authors: | Li, Y, Ren, Y, Li, H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
4N4H
 
 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.1K36me3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.1, ... | | Authors: | Li, Y, Ren, Y, Li, H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
1NDG
 
 | | Crystal structure of Fab fragment of antibody HyHEL-8 complexed with its antigen lysozyme | | Descriptor: | ACETIC ACID, Lysozyme C, antibody kappa light chain, ... | | Authors: | Mariuzza, R.A, Li, Y, Li, H, Yang, F, Smith-Gill, S.J. | | Deposit date: | 2002-12-09 | | Release date: | 2003-06-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray snapshots of the maturation of an antibody response to a protein antigen
Nat.Struct.Biol., 10, 2003
|
|
1NDM
 
 | |
