3Q3C
 
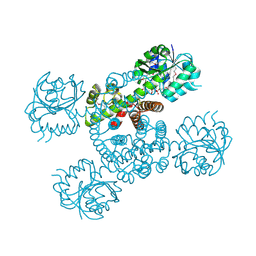 | | Crystal structure of a serine dehydrogenase from Pseudomonas aeruginosa pao1 in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Probable 3-hydroxyisobutyrate dehydrogenase | | Authors: | Tan, K, Singer, A.U, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-12-21 | | Release date: | 2011-02-23 | | Last modified: | 2012-02-01 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Biochemical and Structural Studies of Uncharacterized Protein PA0743 from Pseudomonas aeruginosa Revealed NAD+-dependent L-Serine Dehydrogenase.
J.Biol.Chem., 287, 2012
|
|
3QOK
 
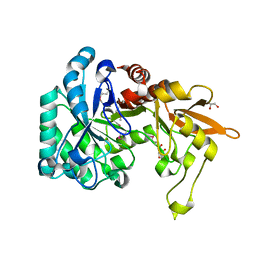 | |
3QOM
 
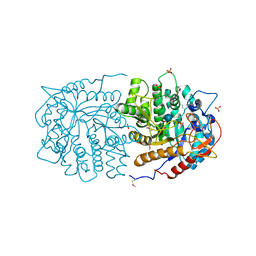 | | Crystal structure of 6-phospho-beta-glucosidase from Lactobacillus plantarum | | Descriptor: | 6-phospho-beta-glucosidase, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Michalska, K, Hatzos-Skintges, C, Bearden, J, Kohler, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-10 | | Release date: | 2011-03-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3QGM
 
 | | p-nitrophenyl phosphatase from Archaeoglobus fulgidus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, p-nitrophenyl phosphatase (Pho2) | | Authors: | Osipiuk, J, Zheng, H, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-01-24 | | Release date: | 2011-02-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | p-nitrophenyl phosphatase from Archaeoglobus fulgidus.
To be Published
|
|
3QSL
 
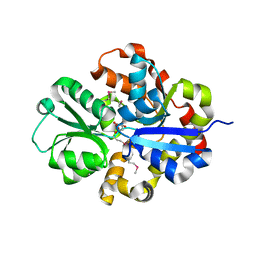 | | Structure of CAE31940 from Bordetella bronchiseptica RB50 | | Descriptor: | CITRIC ACID, Putative exported protein | | Authors: | Bajor, J, Kagan, O, Chruszcz, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-21 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of pyrimidine/thiamin biosynthesis precursor-like domain-containing protein CAE31940 from proteobacterium Bordetella bronchiseptica RB50, and evolutionary insight into the NMT1/THI5 family.
J Struct Funct Genomics, 15, 2014
|
|
3QOD
 
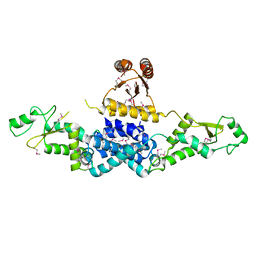 | | Crystal Structure of Heterocyst Differentiation Protein, HetR from Fischerella mv11 | | Descriptor: | Heterocyst differentiation protein | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-09 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structure of transcription factor HetR required for heterocyst differentiation in cyanobacteria.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3QTD
 
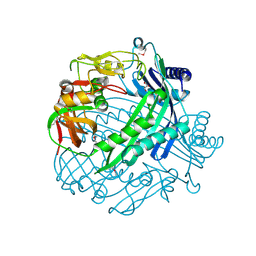 | | Crystal structure of putative modulator of gyrase (PmbA) from Pseudomonas aeruginosa PAO1 | | Descriptor: | GLYCEROL, PmbA protein | | Authors: | Tkaczuk, K.L, Chruszcz, M, Evdokimova, E, Liu, F, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-22 | | Release date: | 2011-03-30 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of putative modulator of gyrase (PmbA) from Pseudomonas aeruginosa PAO1
To be Published
|
|
3QWU
 
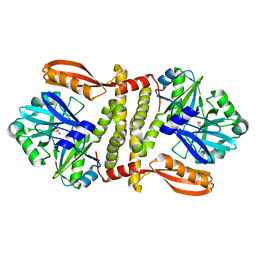 | | Putative ATP-dependent DNA ligase from Aquifex aeolicus. | | Descriptor: | ADENOSINE, CALCIUM ION, DNA ligase, ... | | Authors: | Osipiuk, J, Quartey, P, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-28 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Putative ATP-dependent DNA ligase from Aquifex aeolicus.
To be Published
|
|
3QUF
 
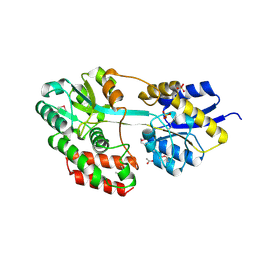 | | The structure of a family 1 extracellular solute-binding protein from Bifidobacterium longum subsp. infantis | | Descriptor: | ACETIC ACID, Extracellular solute-binding protein, family 1, ... | | Authors: | Cuff, M.E, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-23 | | Release date: | 2011-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of a family 1 extracellular solute-binding protein from Bifidobacterium longum subsp. infantis
TO BE PUBLISHED
|
|
3QVQ
 
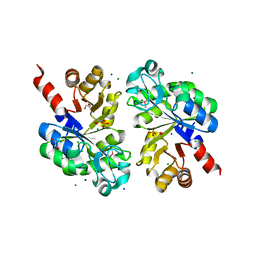 | | The structure of an Oleispira antarctica phosphodiesterase OLEI02445 in complex with the product sn-glycerol-3-phosphate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Kagan, O, Evdokimova, E, Cuff, M.E, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-25 | | Release date: | 2011-04-13 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | The structure of an Oleispira antarctica phosphodiesterase OLEI02445 in complex with the product sn-glycerol-3-phosphate
To be Published
|
|
3QOO
 
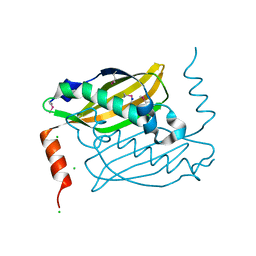 | | Crystal structure of hot-dog-like Taci_0573 protein from Thermanaerovibrio acidaminovorans | | Descriptor: | CHLORIDE ION, SODIUM ION, Uncharacterized protein | | Authors: | Michalska, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-10 | | Release date: | 2011-02-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of hot-dog-like Taci_0573 protein from Thermanaerovibrio acidaminovorans
TO BE PUBLISHED
|
|
3QWT
 
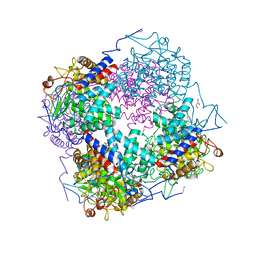 | |
3NZN
 
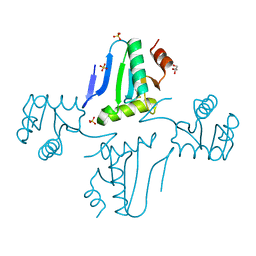 | | The crystal structure of the Glutaredoxin from Methanosarcina mazei Go1 | | Descriptor: | GLYCEROL, Glutaredoxin, SULFATE ION | | Authors: | Zhang, R, Wu, R, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-16 | | Release date: | 2010-07-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The crystal structure of the Glutaredoxin from Methanosarcina mazei Go1
To be Published
|
|
3NYM
 
 | | The crystal structure of functionally unknown protein from Neisseria meningitidis MC58 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Zhang, R, Tan, K, Volkart, L, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-15 | | Release date: | 2010-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of functionally unknown protein from Neisseria meningitidis MC58
To be Published
|
|
3O5Y
 
 | |
3QVO
 
 | | Structure of a Rossmann-fold NAD(P)-binding family protein from Shigella flexneri. | | Descriptor: | 5-MERCAPTO-2-NITRO-BENZOIC ACID, NmrA family protein | | Authors: | Cuff, M.E, Xu, X, Cui, H, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-25 | | Release date: | 2011-06-01 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a Rossmann-fold NAD(P)-binding family protein from Shigella flexneri.
TO BE PUBLISHED
|
|
3QSJ
 
 | | Crystal structure of NUDIX hydrolase from Alicyclobacillus acidocaldarius | | Descriptor: | CALCIUM ION, GLYCEROL, NUDIX hydrolase | | Authors: | Michalska, K, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-21 | | Release date: | 2011-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of NUDIX hydrolase from Alicyclobacillus acidocaldarius
To be Published
|
|
3QTB
 
 | | Structure of the universal stress protein from Archaeoglobus fulgidus in complex with dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, ACETATE ION, Uncharacterized protein | | Authors: | Tkaczuk, K.L, Shumilin, I.A, Chruszcz, M, Cymborowski, M, Xu, X, Di Leo, R, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-22 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional insight into the universal stress protein family.
Evol Appl, 6, 2013
|
|
3QSG
 
 | |
3QXH
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with ADP and 8-aminocaprylic acid | | Descriptor: | 1,2-ETHANEDIOL, 8-aminooctanoic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Porebski, P.J, Klimecka, M.M, Chruszcz, M, Murzyn, K, Minor, C, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QQZ
 
 | | Crystal structure of the C-terminal domain of the yjiK protein from Escherichia coli CFT073 | | Descriptor: | CALCIUM ION, Putative uncharacterized protein yjiK | | Authors: | Stein, A, Chhor, G, Nocek, B, Fenske, R.J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-16 | | Release date: | 2011-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the C-terminal domain of the yjiK protein from Escherichia coli CFT073
TO BE PUBLISHED
|
|
3QXS
 
 | | Crystal structure of dethiobiotin synthetase (BioD) from Helicobacter pylori complexed with ANP | | Descriptor: | 1,2-ETHANEDIOL, Dethiobiotin synthetase, MAGNESIUM ION, ... | | Authors: | Klimecka, M.M, Porebski, P.J, Chruszcz, M, Jablonska, K, Murzyn, K, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural characterization of Helicobacter pylori dethiobiotin synthetase reveals differences between family members.
Febs J., 279, 2012
|
|
3QY3
 
 | | PA2801 protein, a putative Thioesterase from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, Thioesterase | | Authors: | Osipiuk, J, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-16 | | Last modified: | 2012-10-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and activity of the Pseudomonas aeruginosa hotdog-fold thioesterases PA5202 and PA2801.
Biochem.J., 444, 2012
|
|
3NYI
 
 | | The crystal structure of a fat acid (stearic acid)-binding protein from Eubacterium ventriosum ATCC 27560. | | Descriptor: | STEARIC ACID, fat acid-binding protein | | Authors: | Zhang, R, Tan, K, Li, H, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-15 | | Release date: | 2010-09-22 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a fat acid (stearic acid)-binding protein from Eubacterium ventriosum ATCC 27560.
TO BE PUBLISHED
|
|
3NWG
 
 | | The crystal structure of a microcomparments protein from Desulfitobacterium hafniense DCB | | Descriptor: | GLYCEROL, Microcompartments protein | | Authors: | Fan, Y, Volkart, L, Gu, M, Axen, S, Greenleaf, W.B, Kerfeld, C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-09 | | Release date: | 2010-09-22 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a PduT homolog from a novel bacterial microcompartment
To be Published
|
|
