7V7L
 
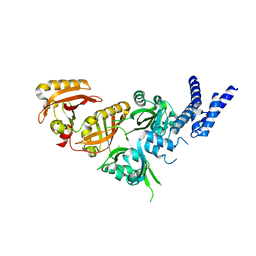 | | Crystal Structure of the Heterodimeric HIF-3a:ARNT Complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Hypoxia-inducible factor 3-alpha | | Authors: | Diao, X, Ren, X, Li, F.W, Sun, X, Wu, D. | | Deposit date: | 2021-08-21 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of oleoylethanolamide as an endogenous ligand for HIF-3 alpha.
Nat Commun, 13, 2022
|
|
8J1F
 
 | | GSK101 bound state of mTRPV4 | | Descriptor: | N-[(2S)-1-{4-[N-(2,4-dichlorobenzene-1-sulfonyl)-L-seryl]piperazin-1-yl}-4-methyl-1-oxopentan-2-yl]-1-benzothiophene-2-carboxamide, Transient receptor potential cation channel subfamily V member 4 | | Authors: | Zhen, W.X, Yang, F. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
8J1H
 
 | | Agonist1 and Ruthenium Red bound state of mTRPV4 | | Descriptor: | 8-fluoranyl-3-[4-(4-fluoranylphenoxy)phenyl]-2-(4-methylpiperazin-1-yl)quinazolin-4-one, Transient receptor potential cation channel subfamily V member 4 | | Authors: | Zhen, W.X, Yang, F. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
8J1D
 
 | | Cryo-EM structure of apo state mTRPV4 | | Descriptor: | Transient receptor potential cation channel subfamily V member 4 | | Authors: | Zhen, W.X, Yang, F. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
8J1B
 
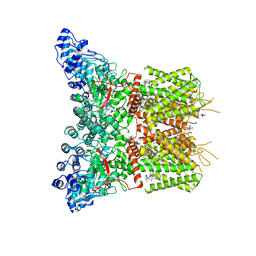 | | GSK101 and Ruthenium Red bound state of mTRPV4 | | Descriptor: | N-[(2S)-1-{4-[N-(2,4-dichlorobenzene-1-sulfonyl)-L-seryl]piperazin-1-yl}-4-methyl-1-oxopentan-2-yl]-1-benzothiophene-2-carboxamide, Transient receptor potential cation channel subfamily V member 4, ruthenium(6+) azanide pentaamino(oxido)ruthenium (1/4/2) | | Authors: | Zhen, W.X, Yang, F. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
8IQZ
 
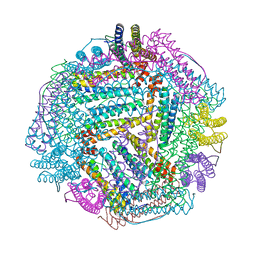 | |
7VNI
 
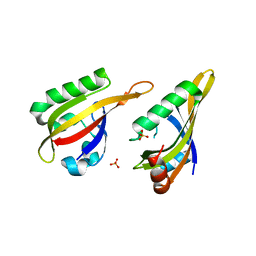 | | AHR-ARNT PAS-B heterodimer | | Descriptor: | Ahr homolog spineless, Aryl hydrocarbon receptor nuclear translocator, SULFATE ION | | Authors: | Dai, S.Y. | | Deposit date: | 2021-10-11 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural insight into the ligand binding mechanism of aryl hydrocarbon receptor.
Nat Commun, 13, 2022
|
|
7VNA
 
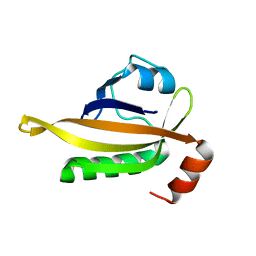 | | drosophlia AHR PAS-B domain | | Descriptor: | Ahr homolog spineless | | Authors: | Dai, S.Y. | | Deposit date: | 2021-10-10 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Structural insight into the ligand binding mechanism of aryl hydrocarbon receptor.
Nat Commun, 13, 2022
|
|
7VNH
 
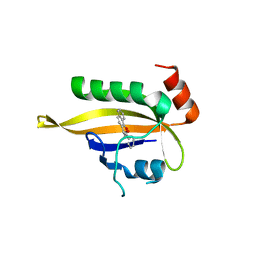 | |
3RJR
 
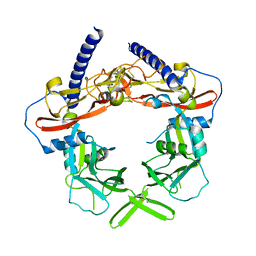 | | Crystal Structure of pro-TGF beta 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Transforming growth factor beta-1 | | Authors: | Zhu, J.H, Shi, M.L, Springer, T.A. | | Deposit date: | 2011-04-15 | | Release date: | 2011-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Latent TGF-Beta structure and activation
Nature, 474, 2011
|
|
7YF5
 
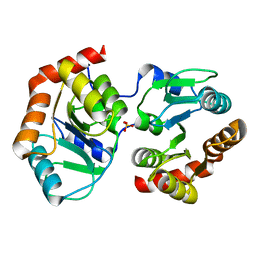 | |
4USG
 
 | | Crystal structure of PC4 W89Y mutant complex with DNA | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP *TP*TP*TP*TP*TP*G)-3', ACTIVATED RNA POLYMERASE II TRANSCRIPTIONAL COACTIVATOR P15 | | Authors: | Zhao, Y, Liu, J. | | Deposit date: | 2014-07-08 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Substitution of Tryptophan 89 with Tyrosine Switches the DNA Binding Mode of Pc4.
Sci.Rep., 5, 2015
|
|
7WJS
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y13157 | | Descriptor: | 2-(2-cyclobutyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-07 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WKY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y13153 | | Descriptor: | 2-(2-cyclopentyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-12 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WMU
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13146 | | Descriptor: | 1,2-ETHANEDIOL, Isoform 4 of Bromodomain-containing protein 2, ~{N}-[4-[2,4-bis(fluoranyl)phenoxy]-3-[2-(2-cyclobutyl-1~{H}-imidazol-5-yl)-5-methyl-4-oxidanylidene-furo[3,2-c]pyridin-7-yl]phenyl]ethanesulfonamide | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-17 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WNI
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13158 | | Descriptor: | 1,2-ETHANEDIOL, 7-[2-[2,4-bis(fluoranyl)phenoxy]-5-(2-oxidanylpropan-2-yl)phenyl]-2-[4-(2-hydroxyethyloxy)-3,5-dimethyl-phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WN5
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13142 | | Descriptor: | 1,2-ETHANEDIOL, Isoform 4 of Bromodomain-containing protein 2, POTASSIUM ION, ... | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-17 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WLN
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13153 | | Descriptor: | 2-(2-cyclopentyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WNA
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13120 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Isoform 4 of Bromodomain-containing protein 2, ... | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-17 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7WMQ
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the inhibitor Y13157 | | Descriptor: | 2-(2-cyclobutyl-1~{H}-imidazol-5-yl)-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, FORMIC ACID, GLYCEROL, ... | | Authors: | Li, J, Zhang, C, Xu, H, Zhuang, X, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-01-16 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-Based Discovery and Optimization of Furo[3,2- c ]pyridin-4(5 H )-one Derivatives as Potent and Second Bromodomain (BD2)-Selective Bromo and Extra Terminal Domain (BET) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7EOK
 
 | | Crystal structure of the Pepper aptamer in complex with HBC485 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[5-[2-(dimethylamino)ethyl-methyl-amino]pyrazin-2-yl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOO
 
 | | Crystal structure of the Pepper aptamer in complex with HBC525 | | Descriptor: | (~{E})-2-(1,3-benzoxazol-2-yl)-3-[4-[2-hydroxyethyl(methyl)amino]phenyl]prop-2-enenitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOP
 
 | | Crystal structure of the Pepper aptamer in complex with HBC620 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[5-[2-hydroxyethyl(methyl)amino]thieno[3,2-b]thiophen-2-yl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOL
 
 | | Crystal structure of the Pepper aptamer in complex with HBC497 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[5-[2-hydroxyethyl(methyl)amino]pyrazin-2-yl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOG
 
 | |
