1UEF
 
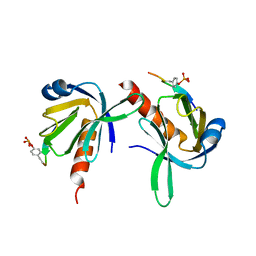 | | Crystal Structure of Dok1 PTB Domain Complex | | Descriptor: | 13-mer peptide from Proto-oncogene tyrosine-protein kinase receptor ret, Docking protein 1 | | Authors: | Shi, N, Ye, S, Liu, Y, Zhou, W, Ding, Y, Lou, Z, Qiang, B, Yan, J, Rao, Z. | | Deposit date: | 2003-05-14 | | Release date: | 2004-05-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Specific Recognition of RET by the Dok1 Phosphotyrosine Binding Domain
J.Biol.Chem., 279, 2004
|
|
4PTC
 
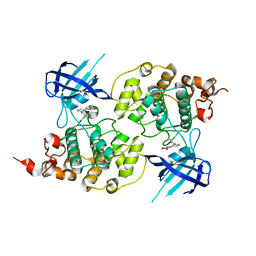 | | Structure of a carboxamide compound (3) (2-{2-[(CYCLOPROPYLCARBONYL)AMINO]PYRIDIN-4-YL}-4-OXO-4H-1LAMBDA~4~,3-THIAZOLE-5-CARBOXAMIDE) to GSK3b | | Descriptor: | 2-[2-(cyclopropylcarbonylamino)pyridin-4-yl]-4-methoxy-1,3-thiazole-5-carboxamide, Glycogen synthase kinase-3 beta | | Authors: | Lewis, H.A, Sivaprakasam, P, Kish, K, Pokross, M, Dubowchik, G.M. | | Deposit date: | 2014-03-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | Discovery of new acylaminopyridines as GSK-3 inhibitors by a structure guided in-depth exploration of chemical space around a pyrrolopyridinone core.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
8H83
 
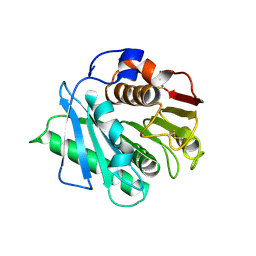 | | Crystal structure of a IsPETase variant V22 from Ideonella sakaiensis | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Wei, H.L, Gao, S.F, Li, Q, Han, X, Gao, J, Liu, W.D. | | Deposit date: | 2022-10-21 | | Release date: | 2024-04-24 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | beta-sheet Engineering of IsPETase for PET Depolymerization
Engineering (Beijing), 2024
|
|
3IOP
 
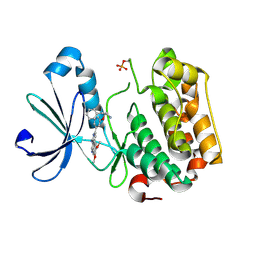 | | PDK-1 in complex with the inhibitor Compound-8i | | Descriptor: | 2-(5-{[(2R)-2-amino-3-phenylpropyl]oxy}pyridin-3-yl)-8,9-dimethoxybenzo[c][2,7]naphthyridin-4-amine, 3-phosphoinositide-dependent protein kinase 1 | | Authors: | Olland, A.M. | | Deposit date: | 2009-08-14 | | Release date: | 2010-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The identification of 8,9-dimethoxy-5-(2-aminoalkoxy-pyridin-3-yl)-benzo[c][2,7]naphthyridin-4-ylamines as potent inhibitors of 3-phosphoinositide-dependent kinase-1 (PDK-1).
Eur.J.Med.Chem., 45, 2010
|
|
4RFY
 
 | | Crystal structure of BTK kinase domain complexed with 6-(dimethylamino)-2-[2-(hydroxymethyl)-3-[1-methyl-5-[[5-(morpholine-4-carbonyl)-2-pyridyl]amino]-6-oxo-3-pyridyl]phenyl]-3,4-dihydroisoquinolin-1-one | | Descriptor: | 6-(dimethylamino)-2-[2-(hydroxymethyl)-3-(1-methyl-5-{[5-(morpholin-4-ylcarbonyl)pyridin-2-yl]amino}-6-oxo-1,6-dihydropyridin-3-yl)phenyl]-3,4-dihydroisoquinolin-1(2H)-one, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Kuglstatter, A, Wong, A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Finding the perfect spot for fluorine: Improving potency up to 40-fold during a rational fluorine scan of a Bruton's Tyrosine Kinase (BTK) inhibitor scaffold.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
8WSP
 
 | | Crystal structure of SFTSV Gn and antibody SF5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ab5-H, Ab5-L, ... | | Authors: | Chang, Z, Gao, F, Wu, Y. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Bispecific antibodies targeting two glycoproteins on SFTSV exhibit synergistic neutralization and protection in a mouse model.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8WSU
 
 | | Crystal structure of SFTSV Gc and antibody | | Descriptor: | Ab-H, Ab-L, Glycoprotein C | | Authors: | Chang, Z, Gao, F, Wu, Y. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Bispecific antibodies targeting two glycoproteins on SFTSV exhibit synergistic neutralization and protection in a mouse model.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8WSN
 
 | | Crystal structure of SFTSV Gn and antibody SF1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ab1-H, ... | | Authors: | Chang, Z, Gao, F, Wu, Y. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bispecific antibodies targeting two glycoproteins on SFTSV exhibit synergistic neutralization and protection in a mouse model.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3ION
 
 | | PDK1 in complex with Compound 8h | | Descriptor: | 2-(5-{[(2S)-2-amino-3-phenylpropyl]oxy}pyridin-3-yl)-8,9-dimethoxybenzo[c][2,7]naphthyridin-4-amine, 3-phosphoinositide-dependent protein kinase 1, SULFATE ION | | Authors: | Olland, A.M. | | Deposit date: | 2009-08-14 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The identification of 8,9-dimethoxy-5-(2-aminoalkoxy-pyridin-3-yl)-benzo[c][2,7]naphthyridin-4-ylamines as potent inhibitors of 3-phosphoinositide-dependent kinase-1 (PDK-1).
Eur.J.Med.Chem., 45, 2010
|
|
4RKW
 
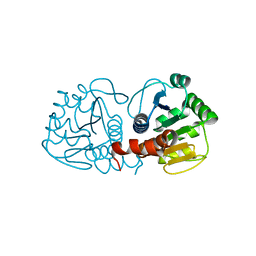 | | Crystal structure of DJ-1 | | Descriptor: | Protein DJ-1 | | Authors: | Liddington, R.C. | | Deposit date: | 2014-10-14 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Transnitrosylation from DJ-1 to PTEN attenuates neuronal cell death in parkinson's disease models.
J.Neurosci., 34, 2014
|
|
9JHP
 
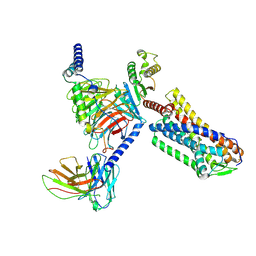 | | Cryo-EM structure of GPR4 complexed with miniG13 in pH6.8 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yue, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-09-10 | | Release date: | 2025-04-23 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis of stepwise proton sensing-mediated GPCR activation.
Cell Res., 35, 2025
|
|
9JFZ
 
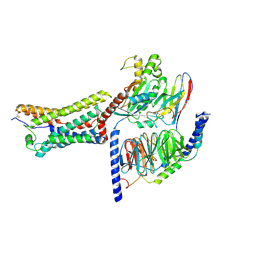 | | Cryo-EM structure of intermediate state GPR4 complexed with miniGs/q in pH7.5 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yue, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-09-05 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of stepwise proton sensing-mediated GPCR activation.
Cell Res., 35, 2025
|
|
9JFX
 
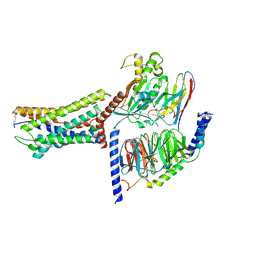 | | Cryo-EM structure of GPR4 complexed with miniGs/q in pH7.5 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yue, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-09-05 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural basis of stepwise proton sensing-mediated GPCR activation.
Cell Res., 35, 2025
|
|
9JFW
 
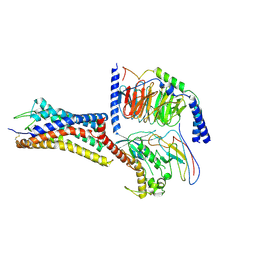 | | Cryo-EM structure of GPR4 complexed with Gs in pH6.8 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yue, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-09-05 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis of stepwise proton sensing-mediated GPCR activation.
Cell Res., 35, 2025
|
|
9JFU
 
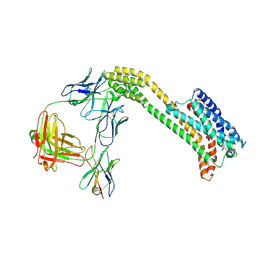 | | Cryo-EM structure of inactive GPR4 with NE52-QQ57 | | Descriptor: | Anti-bril fab light chain, Anti-fab nanobody, G-protein coupled receptor 4,Soluble cytochrome b562, ... | | Authors: | Yue, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-09-05 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural basis of stepwise proton sensing-mediated GPCR activation.
Cell Res., 35, 2025
|
|
9JFV
 
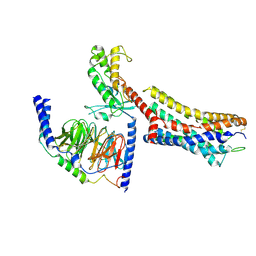 | | Cryo-EM structure of GPR4 complexed with miniGs/q in pH6.8 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yue, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-09-05 | | Release date: | 2025-04-23 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural basis of stepwise proton sensing-mediated GPCR activation.
Cell Res., 35, 2025
|
|
4RFZ
 
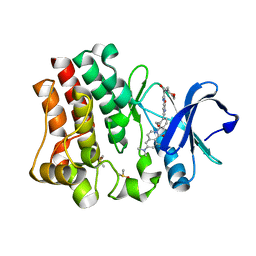 | |
4RG0
 
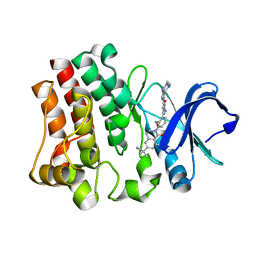 | | Crystal structure of BTK kinase domain complexed with 2-[8-fluoro-2-[2-(hydroxymethyl)-3-[1-methyl-5-[[5-(4-methylpiperazin-1-yl)-2-pyridyl]amino]-6-oxo-3-pyridyl]phenyl]-1-oxo-3,4-dihydroisoquinolin-6-yl]-2-methyl-propanenitrile | | Descriptor: | 2-{8-fluoro-2-[2-(hydroxymethyl)-3-(1-methyl-5-{[5-(4-methylpiperazin-1-yl)pyridin-2-yl]amino}-6-oxo-1,6-dihydropyridin-3-yl)phenyl]-1-oxo-1,2,3,4-tetrahydroisoquinolin-6-yl}-2-methylpropanenitrile, Tyrosine-protein kinase BTK | | Authors: | Kuglstatter, A, Wong, A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Finding the perfect spot for fluorine: Improving potency up to 40-fold during a rational fluorine scan of a Bruton's Tyrosine Kinase (BTK) inhibitor scaffold.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4RKY
 
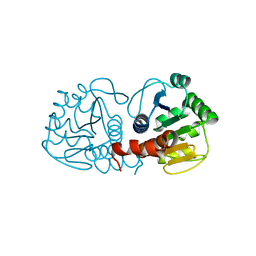 | | Crystal structure of DJ-1 isoform X1 | | Descriptor: | Protein DJ-1 | | Authors: | Liddington, R.C. | | Deposit date: | 2014-10-14 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Transnitrosylation from DJ-1 to PTEN attenuates neuronal cell death in parkinson's disease models.
J.Neurosci., 34, 2014
|
|
3WGJ
 
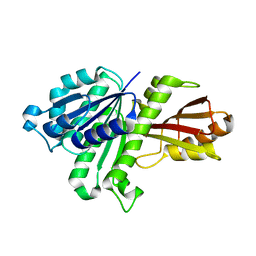 | |
3DI6
 
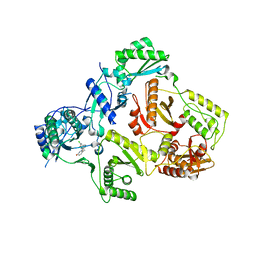 | | HIV-1 RT with pyridazinone non-nucleoside inhibitor | | Descriptor: | 6-(4-chloro-2-fluoro-3-phenoxybenzyl)pyridazin-3(2H)-one, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Harris, S.F, Villasenor, A, Dunten, P. | | Deposit date: | 2008-06-19 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery and optimization of pyridazinone non-nucleoside inhibitors of HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3WGN
 
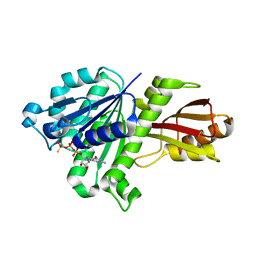 | | STAPHYLOCOCCUS AUREUS FTSZ bound with GTP-gamma-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Cell division protein FtsZ | | Authors: | Matsui, T, Mogi, N, Tanaka, I, Yao, M. | | Deposit date: | 2013-08-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Structural change in FtsZ Induced by intermolecular interactions between bound GTP and the T7 loop
J.Biol.Chem., 289, 2014
|
|
6KIF
 
 | | Structure of cyanobacterial photosystem I-IsiA-flavodoxin supercomplex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Cao, P, Cao, D.F, Si, L, Su, X.D, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex.
Nat.Plants, 6, 2020
|
|
9II2
 
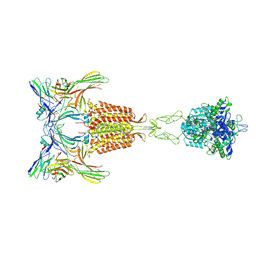 | | Cryo-EM Structure of the 2:2 Complex of mGlu3 and beta-arrestin1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-arrestin-1, CHOLESTEROL, ... | | Authors: | Wen, T.L, Du, M, Yang, X, Shen, Y.Q. | | Deposit date: | 2024-06-18 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis of beta-arrestin coupling to the metabotropic glutamate receptor mGlu3.
Nat.Chem.Biol., 2025
|
|
9II3
 
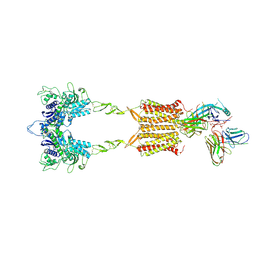 | | Cryo-EM Structure of the 2:1 Complex of mGlu3 and beta-arrestin1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-arrestin-1, CHOLESTEROL, ... | | Authors: | Wen, T.L, Du, M, Yang, X, Shen, Y.Q. | | Deposit date: | 2024-06-19 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular basis of beta-arrestin coupling to the metabotropic glutamate receptor mGlu3.
Nat.Chem.Biol., 2025
|
|
