2EWC
 
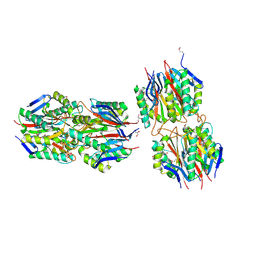 | | Structure of hypothetical protein from Streptococcus pyogenes M1 GAS, member of highly conserved yjgF family of proteins | | Descriptor: | GLYCEROL, conserved hypothetical protein | | Authors: | Nocek, B, Li, H, Clancy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-11-02 | | Release date: | 2005-12-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of hypothetical protein from Streptococcus pyogenes M1 GAS, member of highly conserved yjgF family of proteins
To be Published
|
|
2FB5
 
 | | Structural Genomics; The crystal structure of the hypothetical membrane spanning protein from Bacillus cereus | | Descriptor: | hypothetical Membrane Spanning Protein | | Authors: | Zhang, R, Zhou, M, Ginell, S, Abdullah, J, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-08 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of the hypothetical membrane spanning protein from Bacillus cereus
To be Published
|
|
1RLH
 
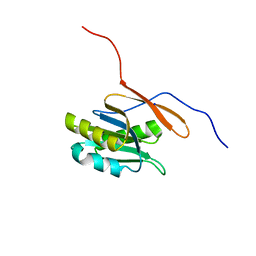 | | Structure of a conserved protein from Thermoplasma acidophilum | | Descriptor: | SODIUM ION, conserved hypothetical protein | | Authors: | Cuff, M.E, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a conserved protein from T. acidophilum
To be Published
|
|
2FDR
 
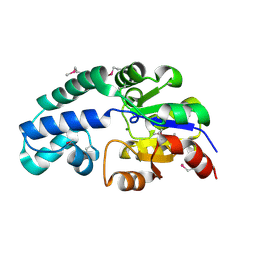 | | Crystal Structure of Conserved Haloacid Dehalogenase-like Protein of Unknown Function ATU0790 from Agrobacterium tumefaciens str. C58 | | Descriptor: | MAGNESIUM ION, conserved hypothetical protein | | Authors: | Nocek, B, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of hydrolases/phosphatases-like fold protein
from Agrobacterium tumefaciens str. C58
To be Published
|
|
2FZV
 
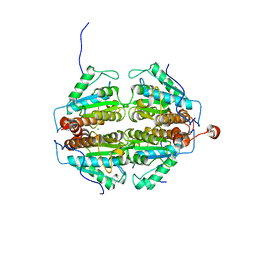 | | Crystal Structure of an apo form of a Flavin-binding Protein from Shigella flexneri | | Descriptor: | CALCIUM ION, CHLORIDE ION, putative arsenical resistance protein | | Authors: | Vorontsov, I.I, Minasov, G, Brunzelle, J.S, Shuvalova, L, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-10 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an apo form of Shigella flexneri ArsH protein with an NADPH-dependent FMN reductase activity
Protein Sci., 16, 2007
|
|
2F6S
 
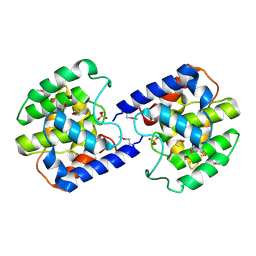 | | Structure of cell filamentation protein (fic) from Helicobacter pylori | | Descriptor: | PHOSPHATE ION, ZINC ION, cell filamentation protein, ... | | Authors: | Cuff, M.E, Xu, X, Zheng, H, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-11-29 | | Release date: | 2006-01-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of cell filamentation protein (fic) from Helicobacter pylori
To be Published
|
|
2F96
 
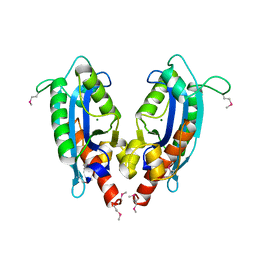 | | 2.1 A crystal structure of Pseudomonas aeruginosa rnase T (Ribonuclease T) | | Descriptor: | MAGNESIUM ION, Ribonuclease T | | Authors: | Zheng, H, Chruszcz, M, Cymborowski, M, Wang, Y, Gorodichtchenskaia, E, Skarina, T, Guthrie, J, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-05 | | Release date: | 2006-02-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of RNase T, an Exoribonuclease Involved in tRNA Maturation and End Turnover.
Structure, 15, 2007
|
|
2F06
 
 | |
2FBL
 
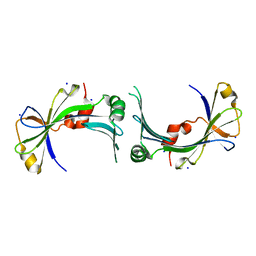 | | The crystal structure of the hypothetical protein NE1496 | | Descriptor: | SODIUM ION, hypothetical protein NE1496 | | Authors: | Lunin, V.V, Skarina, T, Onopriyenko, O, Binkowski, T.A, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-09 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the hypothetical protein NE1496
To be Published
|
|
1O8B
 
 | | Structure of Escherichia coli ribose-5-phosphate isomerase, RpiA, complexed with arabinose-5-phosphate. | | Descriptor: | 5-O-phosphono-beta-D-arabinofuranose, RIBOSE 5-PHOSPHATE ISOMERASE | | Authors: | Zhang, R.-g, Andersson, C.E, Savchenko, A, Skarina, T, Evdokimova, E, Beasley, S, Arrowsmith, C.H, Edwards, A.M, Joachimiak, A, Mowbray, S.L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-11-26 | | Release date: | 2003-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of Escherichia Coli Ribose-5-Phosphate Isomerase: A Ubiquitous Enzyme of the Pentose Phosphate Pathway and the Calvin Cycle
Structure, 11, 2003
|
|
2G39
 
 | | Crystal structure of coenzyme A transferase from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Acetyl-CoA hydrolase | | Authors: | Chang, C, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-17 | | Release date: | 2006-04-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of coenzyme A transferase from Pseudomonas aeruginosa
To be Published
|
|
2G03
 
 | | Structure of a putative cell filamentation protein from Neisseria meningitidis. | | Descriptor: | ACETIC ACID, ISOPROPYL ALCOHOL, hypothetical protein NMA0004 | | Authors: | Cuff, M.E, Bigelow, L, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-10 | | Release date: | 2006-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a putative cell filamentation protein from Neisseria meningitidis.
To be Published
|
|
2GAU
 
 | | Crystal structure of transcriptional regulator, Crp/Fnr family from Porphyromonas gingivalis (APC80792), Structural genomics, MCSG | | Descriptor: | transcriptional regulator, Crp/Fnr family | | Authors: | Rotella, F.J, Zhang, R.G, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-09 | | Release date: | 2006-04-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9-A crystal structure of transcriptional regulator, Crp/Fnr family from Porphyromonas gingivalis
To be Published
|
|
2G7Z
 
 | | Conserved DegV-like Protein of Unknown Function from Streptococcus pyogenes M1 GAS Binds Long-chain Fatty Acids | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, ... | | Authors: | Nocek, B, Volkart, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-01 | | Release date: | 2006-04-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conserved hypothetical protein from Streptococcus pyogenes M1 GAS discloses long-fatty acid (heptadecanoic acid) binding function
To be Published
|
|
2FB0
 
 | | Crystal Structure of Conserved Protein of Unknown Function from Bacteroides thetaiotaomicron VPI-5482 at 2.10 A Resolution, Possible Oxidoreductase | | Descriptor: | ACETATE ION, GLYCEROL, conserved hypothetical protein | | Authors: | Nocek, B, Hatzos, C, Abdullah, J, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-08 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of conserved hypothetical protein from Bacteroides thetaiotaomicron VPI-5482
at 2.10 resolution
To be Published
|
|
2FCK
 
 | | Structure of a putative ribosomal-protein-serine acetyltransferase from Vibrio cholerae. | | Descriptor: | GLYCEROL, NITRATE ION, ribosomal-protein-serine acetyltransferase, ... | | Authors: | Cuff, M.E, Li, H, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-12 | | Release date: | 2006-02-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an acetyltransferase protein from Vibrio cholerae strain N16961.
Proteins, 69, 2007
|
|
1SFS
 
 | | 1.07 A crystal structure of an uncharacterized B. stearothermophilus protein | | Descriptor: | Hypothetical protein, PHOSPHATE ION | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Moy, S.F, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-20 | | Release date: | 2004-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | 1.07 A crystal structure of an uncharacterized B. stearothermophilus protein
To be Published
|
|
2GEN
 
 | | Structural Genomics, the crystal structure of a probable transcriptional regulator from Pseudomonas aeruginosa PAO1 | | Descriptor: | probable transcriptional regulator | | Authors: | Tan, K, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-20 | | Release date: | 2006-04-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of a probable transcriptional regulator from Pseudomonas aeruginosa PAO1
To be Published
|
|
1RXQ
 
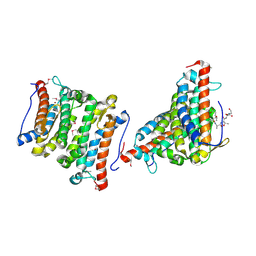 | | YfiT from Bacillus subtilis is a probable metal-dependent hydrolase with an unusual four-helix bundle topology | | Descriptor: | ALANINE, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Rajan, S.S, Yang, X, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-12-18 | | Release date: | 2004-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | YfiT from Bacillus subtilis Is a Probable Metal-Dependent Hydrolase with an Unusual Four-Helix Bundle Topology
Biochemistry, 43, 2004
|
|
1SH8
 
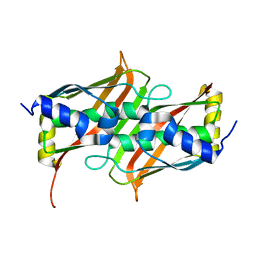 | | 1.5 A Crystal Structure of a Protein of Unknown Function PA5026 from Pseudomonas aeruginosa, Probable Thioesterase | | Descriptor: | hypothetical protein PA5026 | | Authors: | Zhang, R, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-25 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein PA5026 from Pseudomonas aeruginosa
To be Published
|
|
1RYL
 
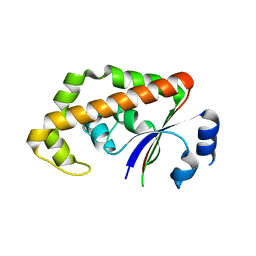 | | The Crystal Structure of a Protein of Unknown Function YfbM from Escherichia coli | | Descriptor: | Hypothetical protein yfbM | | Authors: | Zhang, R, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.6A crystal structure of a hypothetical protein yfbM from E. coli
To be Published
|
|
2G84
 
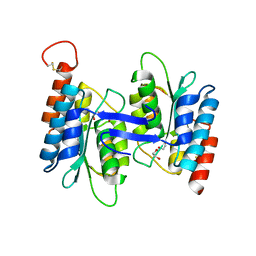 | | Cytidine and deoxycytidylate deaminase zinc-binding region from Nitrosomonas europaea. | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Cytidine and deoxycytidylate deaminase zinc-binding region, ... | | Authors: | Osipiuk, J, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-01 | | Release date: | 2006-04-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of Cytidine and deoxycytidylate deaminase zinc-binding region from Nitrosomonas europaea.
To be Published
|
|
4OVJ
 
 | |
1S5U
 
 | | Crystal Structure of Hypothetical Protein EC709 from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, Protein ybgC, SULFATE ION | | Authors: | Kim, Y, Joachimiak, A, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-01-21 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Hypothetical Protein EC709 from Escherichia coli
To be Published
|
|
1S7I
 
 | | 1.8 A Crystal Structure of a Protein of Unknown Function PA1349 from Pseudomonas aeruginosa | | Descriptor: | hypothetical protein PA1349 | | Authors: | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-01-29 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8A crystal structure of a hypothetical protein PA1349 from Pseudomonas aeruginosa
To be Published
|
|
