1JUH
 
 | | Crystal Structure of Quercetin 2,3-dioxygenase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fusetti, F, Schroeter, K.H, Steiner, R.A, Dijkstra, B.W. | | Deposit date: | 2001-08-24 | | Release date: | 2002-05-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the copper-containing quercetin 2,3-dioxygenase from Aspergillus japonicus.
Structure, 10, 2002
|
|
1CRU
 
 | | SOLUBLE QUINOPROTEIN GLUCOSE DEHYDROGENASE FROM ACINETOBACTER CALCOACETICUS IN COMPLEX WITH PQQ AND METHYLHYDRAZINE | | Descriptor: | CALCIUM ION, GLYCEROL, METHYLHYDRAZINE, ... | | Authors: | Oubrie, A, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1999-08-16 | | Release date: | 2000-03-01 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active-site structure of the soluble quinoprotein glucose dehydrogenase complexed with methylhydrazine: a covalent cofactor-inhibitor complex.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1ORB
 
 | | ACTIVE SITE STRUCTURAL FEATURES FOR CHEMICALLY MODIFIED FORMS OF RHODANESE | | Descriptor: | ACETATE ION, CARBOXYMETHYLATED RHODANESE | | Authors: | Gliubich, F, Gazerro, M, Zanotti, G, Delbono, S, Berni, R. | | Deposit date: | 1995-07-24 | | Release date: | 1995-10-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active site structural features for chemically modified forms of rhodanese.
J.Biol.Chem., 271, 1996
|
|
1PBE
 
 | |
1TCM
 
 | |
1KB0
 
 | |
1LTS
 
 | |
1LTT
 
 | |
1SKZ
 
 | | PROTEASE INHIBITOR | | Descriptor: | ANTISTASIN, CHLORIDE ION | | Authors: | Krengel, U, Dijkstra, B.W. | | Deposit date: | 1997-04-16 | | Release date: | 1997-10-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of antistasin at 1.9 A resolution and its modelled complex with blood coagulation factor Xa.
EMBO J., 16, 1997
|
|
1AG1
 
 | |
1UKR
 
 | | STRUCTURE OF ENDO-1,4-BETA-XYLANASE C | | Descriptor: | ENDO-1,4-B-XYLANASE I | | Authors: | Krengel, U, Dijkstra, B.W. | | Deposit date: | 1996-08-23 | | Release date: | 1997-12-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structure of Endo-1,4-beta-xylanase I from Aspergillus niger: molecular basis for its low pH optimum.
J.Mol.Biol., 263, 1996
|
|
1B48
 
 | |
1EFI
 
 | |
1H1M
 
 | | CRYSTAL STRUCTURE OF QUERCETIN 2,3-DIOXYGENASE ANAEROBICALLY COMPLEXED WITH THE SUBSTRATE KAEMPFEROL | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Steiner, R.A, Dijkstra, B.W. | | Deposit date: | 2002-07-19 | | Release date: | 2002-11-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anaerobic Enzyme.Substrate Structures Provide Insight Into the Reaction Mechanism of the Copper- Dependent Quercetin 2,3-Dioxygenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1H1I
 
 | | CRYSTAL STRUCTURE OF QUERCETIN 2,3-DIOXYGENASE ANAEROBICALLY COMPLEXED WITH THE SUBSTRATE QUERCETN | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Steiner, R.A, Dijkstra, B.W. | | Deposit date: | 2002-07-15 | | Release date: | 2002-11-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Anaerobic Enzyme.Substrate Structures Provide Insight Into the Reaction Mechanism of the Copper- Dependent Quercetin 2,3-Dioxygenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1HTL
 
 | |
2MAD
 
 | |
2WWV
 
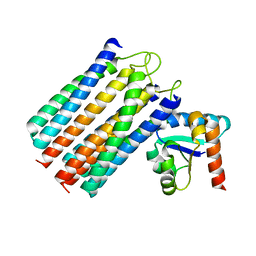 | | NMR structure of the IIAchitobiose-IIBchitobiose complex of the N,N'- diacetylchitoboise brance of the E. coli phosphotransferase system. | | Descriptor: | N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIA COMPONENT, N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIB COMPONENT | | Authors: | Sang, Y.S, Cai, M, Clore, G.M. | | Deposit date: | 2009-10-29 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Iiachitobose-Iibchitobiose Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia Coli Phosphotransfer System
J.Biol.Chem., 285, 2010
|
|
2WY2
 
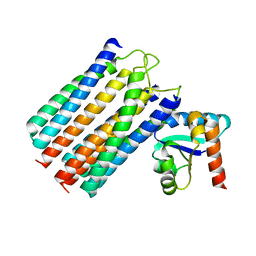 | | NMR structure of the IIAchitobiose-IIBchitobiose phosphoryl transition state complex of the N,N'-diacetylchitoboise brance of the E. coli phosphotransferase system. | | Descriptor: | N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIA COMPONENT, N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIB COMPONENT, PHOSPHITE ION | | Authors: | Sang, Y.S, Cai, M, Clore, G.M. | | Deposit date: | 2009-11-11 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Iiachitobose-Iibchitobiose Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia Coli Phosphotransfer System
J.Biol.Chem., 285, 2010
|
|
3BMV
 
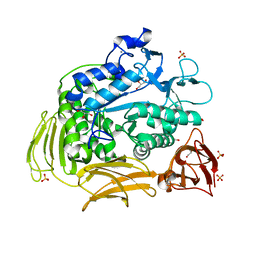 | | Cyclodextrin glycosyl transferase from Thermoanerobacterium thermosulfurigenes EM1 mutant S77P | | Descriptor: | CALCIUM ION, Cyclomaltodextrin glucanotransferase, GLYCEROL, ... | | Authors: | Rozeboom, H.J, van Oosterwijk, N, Dijkstra, B.W. | | Deposit date: | 2007-12-13 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Elimination of competing hydrolysis and coupling side reactions of a cyclodextrin glucanotransferase by directed evolution.
Biochem.J., 413, 2008
|
|
3BMW
 
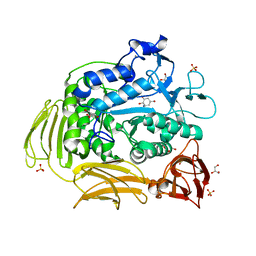 | | Cyclodextrin glycosyl transferase from Thermoanerobacterium thermosulfurigenes EM1 mutant S77P complexed with a maltoheptaose inhibitor | | Descriptor: | 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Rozeboom, H.J, van Oosterwijk, N, Dijkstra, B.W. | | Deposit date: | 2007-12-13 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Elimination of competing hydrolysis and coupling side reactions of a cyclodextrin glucanotransferase by directed evolution.
Biochem.J., 413, 2008
|
|
1QDT
 
 | |
1QDR
 
 | |
1TEC
 
 | | CRYSTALLOGRAPHIC REFINEMENT BY INCORPORATION OF MOLECULAR DYNAMICS. THE THERMOSTABLE SERINE PROTEASE THERMITASE COMPLEXED WITH EGLIN-C | | Descriptor: | CALCIUM ION, EGLIN C, SODIUM ION, ... | | Authors: | Gros, P, Dijkstra, B.W, Hol, W.G.J. | | Deposit date: | 1989-05-24 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic refinement by incorporation of molecular dynamics: thermostable serine protease thermitase complexed with eglin c.
Acta Crystallogr.,Sect.B, 45, 1989
|
|
1HTI
 
 | |
