7F9Z
 
 | | GHRP-6-bound ghrelin receptor in complex with Gq | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, Engineered G-alpha-q, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, Y, Zhuang, Y, Xu, P, Xu, H.E, Jiang, Y. | | Deposit date: | 2021-07-05 | | Release date: | 2021-08-18 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition of an acyl-peptide hormone and activation of ghrelin receptor.
Nat Commun, 12, 2021
|
|
7KQR
 
 | | A 1.89-A resolution substrate-bound crystal structure of heme-dependent tyrosine hydroxylase from S. sclerotialus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Heme-dependent L-tyrosine hydroxylase, ... | | Authors: | Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
7KQS
 
 | | A 1.68-A resolution 3-fluoro-L-tyrosine bound crystal structure of heme-dependent tyrosine hydroxylase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-FLUOROTYROSINE, ... | | Authors: | Wang, Y, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.677 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
7KQU
 
 | | A 1.58-A resolution crystal structure of ferric-hydroperoxo intermediate of L-tyrosine hydroxylase in complex with 3-fluoro-L-tyrosine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-FLUOROTYROSINE, ... | | Authors: | Wang, Y, Davis, I, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
7KQT
 
 | |
8H6R
 
 | | LW Domain of Arabidopsis thaliana TFIIS | | Descriptor: | Transcription elongation factor TFIIS | | Authors: | Wang, Y. | | Deposit date: | 2022-10-18 | | Release date: | 2022-11-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | LW Domain of Arabidopsis thaliana TFIIS
To Be Published
|
|
4XCH
 
 | |
3TDL
 
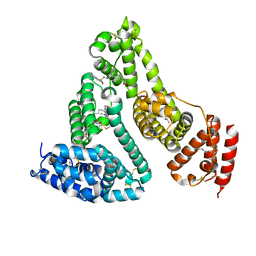 | | Structure of human serum albumin in complex with DAUDA | | Descriptor: | 11-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)undecanoic acid, MYRISTIC ACID, Serum albumin | | Authors: | Wang, Y, Luo, Z, Shi, X, Wang, H, Nie, L. | | Deposit date: | 2011-08-11 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A fluorescent fatty acid probe, DAUDA, selectively displaces two myristates bound in human serum albumin
Protein Sci., 20, 2011
|
|
4FAF
 
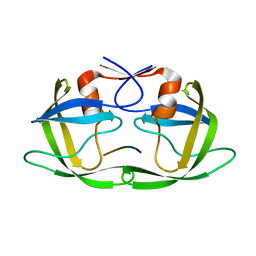 | | Substrate CA/p2 in Complex with a Human Immunodeficiency Virus Type 1 Protease Variant | | Descriptor: | HIV-1 protease, substrate CA/p2 peptide | | Authors: | Wang, Y, Dewdney, T.G, Liu, Z, Reiter, S.J, Brunzelle, J.S, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2012-05-22 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Higher Desolvation Energy Reduces Molecular Recognition in Multi-Drug Resistant HIV-1 Protease.
Biology (Basel), 1, 2012
|
|
2FSZ
 
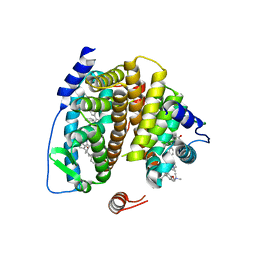 | |
2IMI
 
 | | Structures of an Insect Epsilon-class Glutathione S-transferase from the Malaria Vector Anopheles Gambiae: Evidence for High DDT-detoxifying Activity | | Descriptor: | Epsilon-class Glutathione S-transferase, GLUTATHIONE | | Authors: | Wang, Y, Hemingway, J, Ranson, H, Meehan, E.J, Chen, L. | | Deposit date: | 2006-10-02 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of an insect epsilon class glutathione S-transferase from the malaria vector Anopheles gambiae provides an explanation for the high DDT-detoxifying activity
J.Struct.Biol., 164, 2008
|
|
2GL9
 
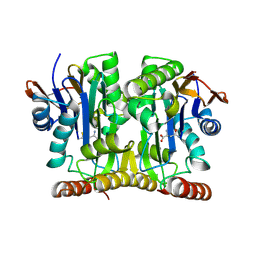 | | Crystal Structure of Glycosylasparaginase-Substrate Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASPARAGINE, Glycosylasparaginase alpha chain, ... | | Authors: | Wang, Y, Guo, H.C. | | Deposit date: | 2006-04-04 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic snapshot of a productive glycosylasparaginase-substrate complex.
J.Mol.Biol., 366, 2007
|
|
2IL3
 
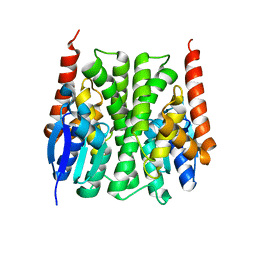 | | Structures of an Insect Epsilon-class Glutathione S-transferase from the Malaria Vector Anopheles Gambiae: Evidence for High DDT-detoxifying Activity | | Descriptor: | Epsilon-class Glutathione S-transferase | | Authors: | Wang, Y, Hemingway, J, Ranson, H, Meehan, E.J, Chen, L. | | Deposit date: | 2006-10-02 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of an insect epsilon class glutathione S-transferase from the malaria vector Anopheles gambiae provides an explanation for the high DDT-detoxifying activity
J.Struct.Biol., 164, 2008
|
|
2IMK
 
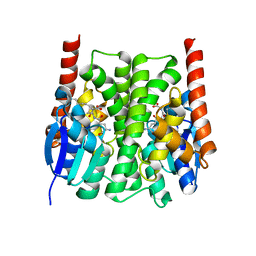 | | Structures of an Insect Epsilon-class Glutathione S-transferase from the Malaria Vector Anopheles Gambiae: Evidence for High DDT-detoxifying Activity | | Descriptor: | Epsilon-class Glutathione S-transferase, S-HEXYLGLUTATHIONE | | Authors: | Wang, Y, Hemingway, J, Ranson, H, Meehan, E.J, Chen, L. | | Deposit date: | 2006-10-02 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of an insect epsilon class glutathione S-transferase from the malaria vector Anopheles gambiae provides an explanation for the high DDT-detoxifying activity
J.Struct.Biol., 164, 2008
|
|
4FAE
 
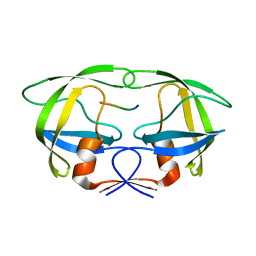 | | Substrate p2/NC in Complex with a Human Immunodeficiency Virus Type 1 Protease Variant | | Descriptor: | HIV-1 protease, Substrate p2/NC peptide | | Authors: | Wang, Y, Dewdney, T.G, Liu, Z, Reiter, S.J, Brunzelle, J.S, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2012-05-22 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Higher Desolvation Energy Reduces Molecular Recognition in Multi-Drug Resistant HIV-1 Protease.
Biology (Basel), 1, 2012
|
|
5X6D
 
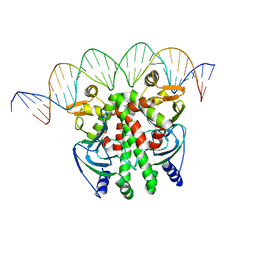 | | Crystal structure of PrfA-DNA binary complex | | Descriptor: | DNA (28-MER), DNA (29-MER), Listeriolysin positive regulatory factor A | | Authors: | Wang, Y, Feng, H, Zhu, Y.L, Gao, P. | | Deposit date: | 2017-02-21 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structural insights into glutathione-mediated activation of the master regulator PrfA in Listeria monocytogenes
Protein Cell, 8, 2017
|
|
2K48
 
 | | NMR Structure of the N-terminal Coiled Coil Domain of the Andes Hantavirus Nucleocapsid Protein | | Descriptor: | Nucleoprotein | | Authors: | Wang, Y, Boudreaux, D.M, Estrada, D.F, Egan, C.W, St Jeor, S.C, De Guzman, R.N. | | Deposit date: | 2008-05-30 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the N-terminal Coiled Coil Domain of the Andes Hantavirus Nucleocapsid Protein.
J.Biol.Chem., 283, 2008
|
|
5X6E
 
 | | Crystal structure of PrfA-DNA binary complex | | Descriptor: | DNA (28-MER), DNA (29-MER), GLUTATHIONE, ... | | Authors: | Wang, Y, Feng, H, Zhu, Y.L, Gao, P. | | Deposit date: | 2017-02-21 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural insights into glutathione-mediated activation of the master regulator PrfA in Listeria monocytogenes
Protein Cell, 8, 2017
|
|
2H52
 
 | |
4IW1
 
 | | HSA-fructose complex | | Descriptor: | D-fructose, PHOSPHATE ION, Serum albumin, ... | | Authors: | Wang, Y, Yu, H, Shi, X, Huang, M. | | Deposit date: | 2013-01-23 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural mechanism of ring-opening reaction of glucose by human serum albumin
J.Biol.Chem., 288, 2013
|
|
4JFJ
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with compound (1S,6R)-10-(1,3-benzothiazol-6-ylsulfonyl)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,6R)-10-(1,3-benzothiazol-6-ylsulfonyl)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4JFM
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with 2-(3,4-dimethoxyphenoxy)ethyl (2S)-1-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]piperidine-2-carboxylate | | Descriptor: | 2-(3,4-dimethoxyphenoxy)ethyl (2S)-1-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4K2C
 
 | | HSA Ligand Free | | Descriptor: | Serum albumin | | Authors: | Wang, Y, Luo, Z, Shi, X, Huang, M. | | Deposit date: | 2013-04-08 | | Release date: | 2013-05-01 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structural mechanism of ring-opening reaction of glucose by human serum albumin.
J. Biol. Chem., 288, 2013
|
|
4JFL
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with 6-({(1S,5R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-2-oxo-3,9-diazabicyclo[3.3.1]non-9-yl}sulfonyl)-1,3-benzothiazol-2(3H)-one | | Descriptor: | 6-({(1S,5R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-2-oxo-3,9-diazabicyclo[3.3.1]non-9-yl}sulfonyl)-1,3-benzothiazol-2(3H)-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4JFI
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with compound 1-[(9S,13R,13aR)-1,3-dimethoxy-8-oxo-5,8,9,10,11,12,13,13a-octahydro-6H-9,13-epiminoazocino[2,1-a]isoquinolin-14-yl]-2-(3,4,5-trimethoxyphenyl)ethane-1,2-dione | | Descriptor: | 1-[(9S,13R,13aR)-1,3-dimethoxy-8-oxo-5,8,9,10,11,12,13,13a-octahydro-6H-9,13-epiminoazocino[2,1-a]isoquinolin-14-yl]-2-(3,4,5-trimethoxyphenyl)ethane-1,2-dione, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
