6TNA
 
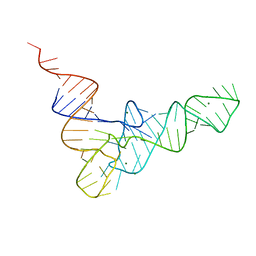 | | CRYSTAL STRUCTURE OF YEAST PHENYLALANINE T-RNA. I.CRYSTALLOGRAPHIC REFINEMENT | | Descriptor: | MAGNESIUM ION, TRNAPHE | | Authors: | Sussman, J.L, Holbrook, S.R, Warrant, R.W, Church, G.M, Kim, S.-H. | | Deposit date: | 1978-11-16 | | Release date: | 1979-01-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement.
J.Mol.Biol., 123, 1978
|
|
3EYI
 
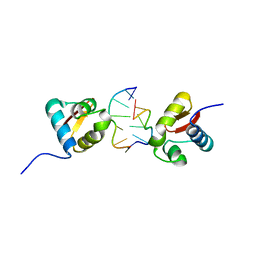 | |
3F23
 
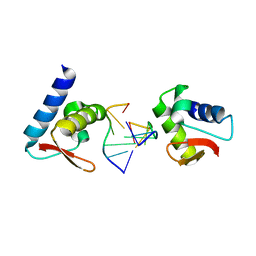 | | Crystal structure of Zalpha in complex with d(CGGCCG) | | Descriptor: | DNA (5'-D(*DTP*DCP*DGP*DGP*DCP*DCP*DG)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Ha, S.C, Choi, J, Kim, K.K. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structures of non-CG-repeat Z-DNAs co-crystallized with the Z-DNA-binding domain, hZ{alpha}ADAR1
Nucleic Acids Res., 37, 2009
|
|
3F22
 
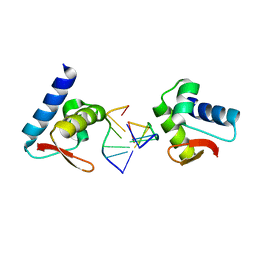 | | Crystal structure of Zalpha in complex with d(CGTACG) | | Descriptor: | DNA (5'-D(*DTP*DCP*DGP*DTP*DAP*DCP*DG)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Ha, S.C, Choi, J, Kim, K.K. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structures of non-CG-repeat Z-DNAs co-crystallized with the Z-DNA-binding domain, hZ{alpha}ADAR1
Nucleic Acids Res., 37, 2009
|
|
3F21
 
 | | Crystal structure of Zalpha in complex with d(CACGTG) | | Descriptor: | DNA (5'-D(*DTP*DCP*DAP*DCP*DGP*DTP*DG)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Ha, S.C, Choi, J, Kim, K.K. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structures of non-CG-repeat Z-DNAs co-crystallized with the Z-DNA-binding domain, hZ{alpha}ADAR1
Nucleic Acids Res., 37, 2009
|
|
1LAP
 
 | | MOLECULAR STRUCTURE OF LEUCINE AMINOPEPTIDASE AT 2.7-ANGSTROMS RESOLUTION | | Descriptor: | Cytosol aminopeptidase, ZINC ION | | Authors: | Burley, S.K, David, P.R, Taylor, A, Lipscomb, W.N. | | Deposit date: | 1990-08-01 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular structure of leucine aminopeptidase at 2.7-A resolution.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
482D
 
 | | RELEASE OF THE CYANO MOIETY IN THE CRYSTAL STRUCTURE OF N-CYANOMETHYL-N-(2-METHOXYETHYL)-DAUNOMYCIN COMPLEXED WITH D(CGATCG) | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*G)-3', N-HYDROXYMETHYL-N-(2-METHOXYETHYL)-DAUNOMYCIN | | Authors: | Saminadin, P, Dautant, A, Mondon, M, Langlois D'Estaintot, B, Courseille, C, Precigoux, G. | | Deposit date: | 1999-07-27 | | Release date: | 1999-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Release of the cyano moiety in the crystal structure of N-cyanomethyl-N-(2-methoxyethyl)-daunomycin complexed with d(CGATCG).
Eur.J.Biochem., 267, 2000
|
|
1D39
 
 | | COVALENT MODIFICATION OF GUANINE BASES IN DOUBLE STRANDED DNA: THE 1.2 ANGSTROMS Z-DNA STRUCTURE OF D(CGCGCG) IN THE PRESENCE OF CUCL2 | | Descriptor: | COPPER (II) ION, DNA (5'-D(*CP*(CU)GP*CP*(CU)GP*CP*(CU)G)-3'), SODIUM ION | | Authors: | Kagawa, T.F, Geierstanger, B.H, Wang, A.H.-J, Ho, P.S. | | Deposit date: | 1991-05-07 | | Release date: | 1992-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Covalent modification of guanine bases in double-stranded DNA. The 1.2-A Z-DNA structure of d(CGCGCG) in the presence of CuCl2.
J.Biol.Chem., 266, 1991
|
|
2F8W
 
 | | Crystal structure of d(CACGTG)2 | | Descriptor: | 1,3-DIAMINOPROPANE, 5'-D(*CP*AP*CP*GP*TP*G)-3', SPERMINE | | Authors: | Narayana, N, Shamala, N, Ganesh, K.N, Viswamitra, M.A. | | Deposit date: | 2005-12-04 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Interaction between the Z-Type DNA Duplex and 1,3-Propanediamine: Crystal Structure of d(CACGTG)2 at 1.2 A Resolution
Biochemistry, 45, 2006
|
|
1NAB
 
 | | The crystal structure of the complex between a disaccharide anthracycline and the DNA hexamer d(CGATCG) reveals two different binding sites involving two DNA duplexes | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*G)-3', 7-[5-(4-AMINO-5-HYDROXY-6-METHYL-TETRAHYDRO-PYRAN-2-YLOXY)-4-HYDROXY-6-METHYL-TETRAHYDRO-PYRAN-2-YLOXY]-6,9,11-TRIHYDROXY-9-(2-HYDROXY-ACETYL)-7,8,9,10-TETRAHYDRO-NAPHTHACENE-5,12-DIONE | | Authors: | Temperini, C, Messori, L, Orioli, P, Di Bugno, C, Animati, F, Ughetto, G. | | Deposit date: | 2002-11-27 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of the complex between a disaccharide anthracycline and the DNA hexamer d(CGATCG) reveals two different binding sites involving two DNA duplexes
Nucleic Acids Res., 31, 2003
|
|
187D
 
 | |
189D
 
 | |
1JES
 
 | | Crystal Structure of a Copper-Mediated Base Pair in DNA | | Descriptor: | 5'-D(*CP*GP*CP*GP*(DPY)P*AP*TP*(DRP)P*CP*GP*CP*G)-3', COPPER (II) ION | | Authors: | Atwell, S, Meggers, E, Spraggon, G, Schultz, P.G. | | Deposit date: | 2001-06-18 | | Release date: | 2001-11-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a Copper-Mediated Base Pair in DNA
J.Am.Chem.Soc., 123, 2001
|
|
263D
 
 | | ISOHELICITY AND PHASING IN DRUG-DNA SEQUENCE RECOGNITION: CRYSTAL STRUCTURE OF A TRIS(BENZIMIDAZOLE)-OLIGONUCLEOTIDE COMPLEX | | Descriptor: | 2''-(4-METHOXYPHENYL)-5-(3-AMINO-1-PYRROLIDINYL)-2,5',2',5''-TRI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Clark, G.R, Gray, E.J, Neidle, S, Li, Y.-H, Leupin, W. | | Deposit date: | 1996-09-27 | | Release date: | 1996-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isohelicity and phasing in drug--DNA sequence recognition: crystal structure of a tris(benzimidazole)--oligonucleotide complex.
Biochemistry, 35, 1996
|
|
145D
 
 | |
7AKR
 
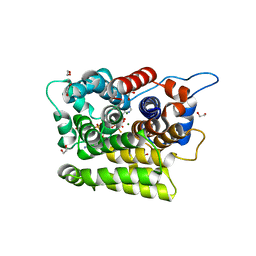 | |
7AKS
 
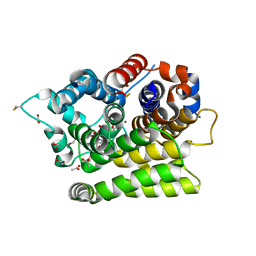 | |
7AZT
 
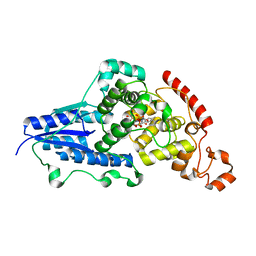 | | X-ray crystallographic structure of (6-4)photolyase from Drosophila melanogaster at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, RE11660p | | Authors: | Cellini, A, Wahlgren, W.Y, Henry, L, Westenhoff, S, Pandey, S. | | Deposit date: | 2020-11-17 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The three-dimensional structure of Drosophila melanogaster (6-4) photolyase at room temperature.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7AYV
 
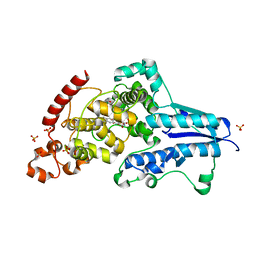 | | X-ray crystallographic structure of (6-4)photolyase from Drosophila melanogaster at cryogenic temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, RE11660p, ... | | Authors: | Cellini, A, Wahlgren, W.Y, Henry, L, Westenhoff, S. | | Deposit date: | 2020-11-13 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The three-dimensional structure of Drosophila melanogaster (6-4) photolyase at room temperature.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
2L4M
 
 | |
242D
 
 | | MAD PHASING STRATEGIES EXPLORED WITH A BROMINATED OLIGONUCLEOTIDE CRYSTAL AT 1.65 A RESOLUTION. | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(CBR)P*G)-3') | | Authors: | Peterson, M.R, Harrop, S.J, McSweeney, S.M, Leonard, G.A, Thompson, A.W, Hunter, W.N, Helliwell, J.R. | | Deposit date: | 1996-06-20 | | Release date: | 1996-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | MAD Phasing Strategies Explored with a Brominated Oligonucleotide Crystal at 1.65A Resolution.
J.Synchrotron Radiat., 3, 1996
|
|
7AQM
 
 | | ADP-ribosylserine hydrolase ARH3 of Latimeria chalumnae in complex with alpha-1''-O-methyl-ADP-ribose (meADPr) | | Descriptor: | ADP-ribosylhydrolase like 2, Adenosine 5'-diphosphoric acid beta-[(3beta,4beta-dihydroxy-5beta-methoxytetrahydrofuran-2alpha-yl)methyl] estere, MAGNESIUM ION | | Authors: | Rack, J.G.M, Zorzini, V, Ahel, I. | | Deposit date: | 2020-10-22 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic insights into the three steps of poly(ADP-ribosylation) reversal.
Nat Commun, 12, 2021
|
|
7ARW
 
 | | Structure of human ARH3 E41A bound to alpha-NAD+ and magnesium | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADP-ribose glycohydrolase ARH3, ... | | Authors: | Rack, J.G.M, Zorzini, V, Ahel, I. | | Deposit date: | 2020-10-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Mechanistic insights into the three steps of poly(ADP-ribosylation) reversal.
Nat Commun, 12, 2021
|
|
5I35
 
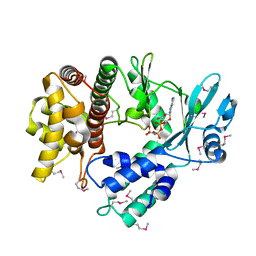 | | Structure of the Human mitochondrial kinase COQ8A R611K with AMPPNP (Cerebellar Ataxia and Ubiquinone Deficiency Through Loss of Unorthodox Kinase Activity) | | Descriptor: | Atypical kinase ADCK3, mitochondrial, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Bingman, C.A, Stefely, J.A, Pagliarini, D.J, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2016-02-09 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cerebellar Ataxia and Coenzyme Q Deficiency through Loss of Unorthodox Kinase Activity.
Mol.Cell, 63, 2016
|
|
4TRA
 
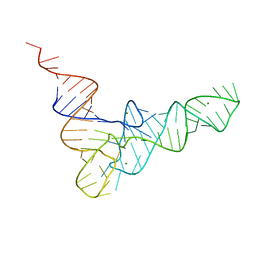 | |
