2IZZ
 
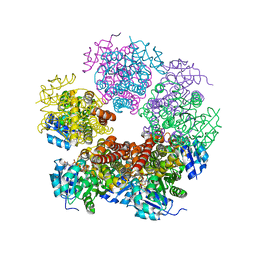 | | Crystal structure of human pyrroline-5-carboxylate reductase | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYRROLINE-5-CARBOXYLATE REDUCTASE 1 | | Authors: | Pike, A.C.W, Guo, K, Kavanagh, K, Pilka, E.S, Berridge, G, Colebrook, S, Bray, J, Salah, E, Savitsky, P, Papagrigoriou, E, Turnbull, A.P, von Delft, F, Arrowsmith, C, Edwards, A, Weigelt, J, Sundstrom, M, Oppermann, U. | | Deposit date: | 2006-07-31 | | Release date: | 2006-08-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Human Pyrroline-5-Carboxylate Reductase
To be Published
|
|
2IXS
 
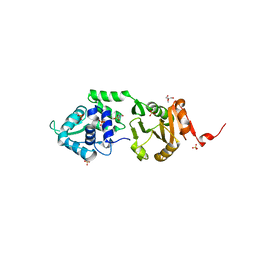 | | Structure of SdaI restriction endonuclease | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SDAI RESTRICTION ENDONUCLEASE, ... | | Authors: | Tamulaitiene, G, Jakubauskas, A, Urbanke, C, Huber, R, Grazulis, S, Siksnys, V. | | Deposit date: | 2006-07-11 | | Release date: | 2006-09-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Rare-Cutting Restriction Enzyme Sdai Reveals Unexpected Domain Architecture
Structure, 14, 2006
|
|
2J6B
 
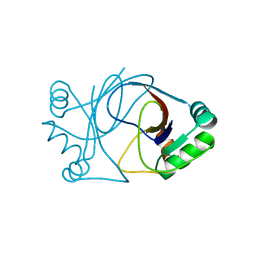 | | crystal structure of AFV3-109, a highly conserved protein from crenarchaeal viruses | | Descriptor: | AFV3-109 | | Authors: | Keller, J, Leulliot, N, Cambillau, C, Campanacci, V, Porciero, S, Prangishvili, D, Cortez, D, Quevillon-Cheruel, S, Van Tilbeurgh, H. | | Deposit date: | 2006-09-27 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Afv3-109, a Highly Conserved Protein from Crenarchaeal Viruses.
Virol J., 4, 2007
|
|
8I7S
 
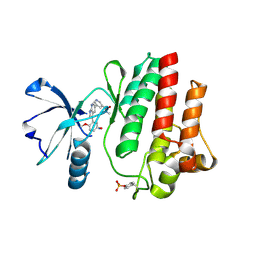 | | The crystal structure of human abl1 kinase domain in complex with ABL1-B1 | | Descriptor: | 5-[3-(2-methoxy-5-oxidanyl-phenyl)-1H-pyrrolo[2,3-b]pyridin-5-yl]-N,N-dimethyl-pyridine-3-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Zhu, C, Zhang, Z. | | Deposit date: | 2023-02-02 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94635463 Å) | | Cite: | Rationally designed BCR-ABL kinase inhibitors for improved leukemia treatment via covalent and pro-/dual-drug targeting strategies.
J Adv Res, 2024
|
|
8I7Z
 
 | | The crystal structure of human abl1 kinase domain in complex with ABL1-B5 | | Descriptor: | 5-[3-(2-methoxy-5-oxidanyl-phenyl)-1H-pyrrolo[2,3-b]pyridin-5-yl]-N,N-dimethyl-pyridine-3-carboxamide, BETA-ALANINE, Tyrosine-protein kinase ABL1 | | Authors: | Zhu, C, Zhang, Z. | | Deposit date: | 2023-02-02 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25495815 Å) | | Cite: | Rationally designed BCR-ABL kinase inhibitors for improved leukemia treatment via covalent and pro-/dual-drug targeting strategies.
J Adv Res, 2024
|
|
8I7T
 
 | | The crystal structure of human abl1 kinase domain in complex with ABL1-B4 | | Descriptor: | Tyrosine-protein kinase ABL1, [3-[5-[5-(dimethylcarbamoyl)pyridin-3-yl]-1H-pyrrolo[2,3-b]pyridin-3-yl]-4-methoxy-phenyl] ethanesulfonate | | Authors: | Zhu, C, Zhang, Z. | | Deposit date: | 2023-02-02 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.80004954 Å) | | Cite: | Rationally designed BCR-ABL kinase inhibitors for improved leukemia treatment via covalent and pro-/dual-drug targeting strategies.
J Adv Res, 2024
|
|
2J9P
 
 | | Crystal structure of the Bacillus subtilis PBP4a, and its complex with a peptidoglycan mimetic peptide. | | Descriptor: | (2R)-2-AMINO-7-{[(1R)-1-CARBOXYETHYL]AMINO}-7-OXOHEPTANOIC ACID, D-ALANINE, D-alanyl-D-alanine carboxypeptidase DacC | | Authors: | Sauvage, E, Herman, R, Kerff, F, Duez, C, Charlier, P. | | Deposit date: | 2006-11-15 | | Release date: | 2007-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Bacillus subtilis penicillin-binding protein 4a, and its complex with a peptidoglycan mimetic peptide.
J. Mol. Biol., 371, 2007
|
|
2IKK
 
 | | Structural Genomics, the crystal structure of the C-terminal domain of Yurk from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | Hypothetical transcriptional regulator yurK, SULFATE ION | | Authors: | Tan, K, Hatzos, C, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-02 | | Release date: | 2006-10-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the C-terminal domain of Yurk from Bacillus subtilis subsp. subtilis str. 168
To be Published
|
|
2IDK
 
 | | Crystal Structure of Rat Glycine N-Methyltransferase Complexed With Folate | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, Glycine N-methyltransferase | | Authors: | Luka, Z, Pakhomova, S, Loukachevitch, L.V, Egli, M, Newcomer, M.E, Wagner, C. | | Deposit date: | 2006-09-15 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 5-methyltetrahydrofolate is bound in intersubunit areas of rat liver folate-binding protein glycine N-methyltransferase.
J.Biol.Chem., 282, 2007
|
|
8IQY
 
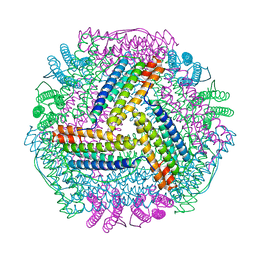 | |
8IQV
 
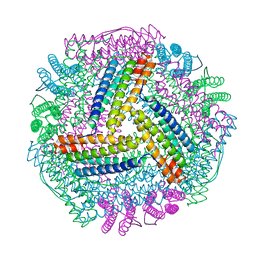 | |
2IQI
 
 | | Crystal structure of protein XCC0632 from Xanthomonas campestris, Pfam DUF330 | | Descriptor: | Hypothetical protein XCC0632 | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Mckenzie, C, Pelletier, L, Wasserman, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-13 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of hypothetical protein XCC0632 from Xanthomonas campestris pv. campestris
To be Published
|
|
8IQW
 
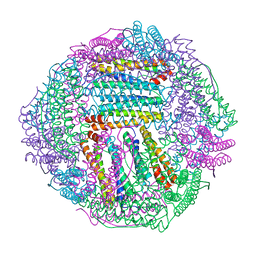 | |
8IQZ
 
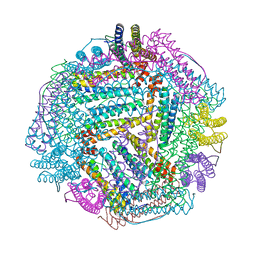 | |
8IY7
 
 | |
8IZZ
 
 | |
2IVF
 
 | | Ethylbenzene dehydrogenase from Aromatoleum aromaticum | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ACETATE ION, ... | | Authors: | Kloer, D.P, Hagel, C, Heider, J, Schulz, G.E. | | Deposit date: | 2006-06-13 | | Release date: | 2006-09-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of Ethylbenzene Dehydrogenase from Aromatoleum Aromaticum
Structure, 14, 2006
|
|
2J0G
 
 | | L-ficolin complexed to N-acetyl-mannosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-mannopyranose, CALCIUM ION, FICOLIN-2, ... | | Authors: | Garlatti, V, Gaboriaud, C. | | Deposit date: | 2006-08-03 | | Release date: | 2007-01-23 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Insights Into the Innate Immune Recognition Specificities of L- and H-Ficolins.
Embo J., 26, 2007
|
|
2J86
 
 | | Structural analysis of the PP2C Family Phosphatase tPphA of Thermosynechococcus elongatus | | Descriptor: | MAGNESIUM ION, PROTEIN SERINE-THREONINE PHOSPHATASE | | Authors: | Schlicker, C, Kloft, N, Forchhammer, K, Becker, S. | | Deposit date: | 2006-10-19 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural Analysis of the Pp2C Phosphatase Tppha from Thermosynechococcus Elongatus: A Flexible Flap Subdomain Controls Access to the Catalytic Site.
J.Mol.Biol., 376, 2008
|
|
8GCC
 
 | | T. cruzi topoisomerase II alpha bound to dsDNA and the covalent inhibitor CT1 | | Descriptor: | 2-{3-[(Z)-iminomethyl]-1H-1,2,4-triazol-1-yl}-1-{(3M)-3-[2-(trifluoromethyl)phenyl]-6H-pyrrolo[3,4-b]pyridin-6-yl}ethan-1-one, DNA (28-MER), DNA topoisomerase 2 | | Authors: | Schenk, A, Deniston, C, Noeske, J. | | Deposit date: | 2023-03-01 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cyanotriazoles are selective topoisomerase II poisons that rapidly cure trypanosome infections.
Science, 380, 2023
|
|
2ID2
 
 | | GAPN T244S mutant X-ray structure at 2.5 A | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Pailot, A, D'Ambrosio, K, Corbier, C, Talfournier, F, Branlant, G. | | Deposit date: | 2006-09-14 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Invariant Thr(244) is essential for the efficient acylation step of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase from Streptococcus mutans.
Biochem.J., 400, 2006
|
|
2IZS
 
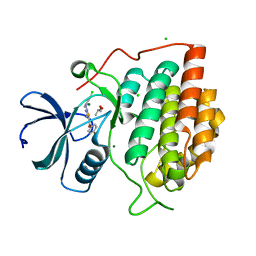 | | Structure of casein kinase gamma 3 in complex with inhibitor | | Descriptor: | CASEIN KINASE I ISOFORM GAMMA-3, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bunkoczi, G, Salah, E, Rellos, P, Das, S, Fedorov, O, Savitsky, P, Debreczeni, J.E, Gileadi, O, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, von Delft, F, Knapp, S. | | Deposit date: | 2006-07-26 | | Release date: | 2006-08-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Inhibitor Binding by Casein Kinases
To be Published
|
|
7B83
 
 | | Structure of SARS-CoV-2 Main Protease bound to pyrithione zinc | | Descriptor: | 3C-like proteinase, 9-oxa-7-thia-1-azonia-8$l^{2}-zincabicyclo[4.3.0]nona-1,3,5-triene, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6Q0Q
 
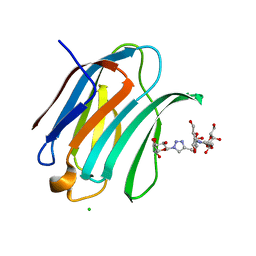 | | Crystal structure of Human galectin-3 CRD in complex with Methyl 3-O-(1-{3-O-[1-(b-D-galactopyranosyl)-1,2,3-triazol-4-yl]-methyl-b-D-galactopyranosyl}-1,2,3-triazol-4-yl)-methyl-b-D-galactopyranoside | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-[4-[[(2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-[4-[[(2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-methoxy-3,5-bis(oxidanyl)oxan-4-yl]oxymethyl]-1,2,3-triazol-1-yl]-3,5-bis(oxidanyl)oxan-4-yl]oxymethyl]-1,2,3-triazol-1-yl]oxane-3,4,5-triol, CHLORIDE ION, Galectin-3 | | Authors: | Kishor, C, Blanchard, H. | | Deposit date: | 2019-08-02 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98601174 Å) | | Cite: | Linear triazole-linked pseudo oligogalactosides as scaffolds for galectin inhibitor development.
Chem.Biol.Drug Des., 96, 2020
|
|
6Q17
 
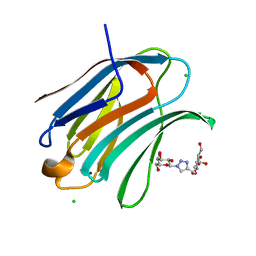 | | Crystal structure of Human galectin-3 CRD in complex with Methyl 3-O-[1-(b-D-galactopyranosyl)-1,2,3-triazol-4-yl]-methyl-b-D-galactopyranoside | | Descriptor: | CHLORIDE ION, Galectin-3, methyl 3-O-[(1-beta-D-galactopyranosyl-1H-1,2,3-triazol-4-yl)methyl]-beta-D-galactopyranoside | | Authors: | Kishor, C, Blanchard, H. | | Deposit date: | 2019-08-02 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Linear triazole-linked pseudo oligogalactosides as scaffolds for galectin inhibitor development.
Chem.Biol.Drug Des., 96, 2020
|
|
