1A6C
 
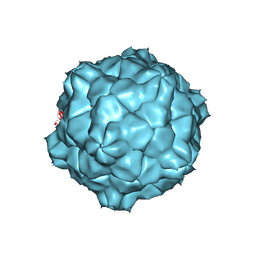 | | STRUCTURE OF TOBACCO RINGSPOT VIRUS | | Descriptor: | TOBACCO RINGSPOT VIRUS CAPSID PROTEIN | | Authors: | Johnson, J.E, Chandrasekar, V. | | Deposit date: | 1998-02-23 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of tobacco ringspot virus: a link in the evolution of icosahedral capsids in the picornavirus superfamily.
Structure, 6, 1998
|
|
5W0Q
 
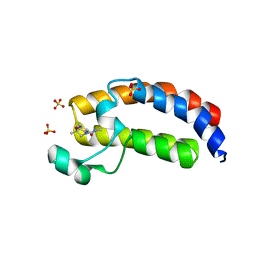 | | CREBBP Bromodomain in complex with Cpd17 (N,2,7-trimethyl-2,3-dihydro-4H-benzo[b][1,4]oxazine-4-carboxamide) | | Descriptor: | (2R)-N,2,7-trimethyl-2,3-dihydro-4H-1,4-benzoxazine-4-carboxamide, CREB-binding protein, SULFATE ION | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-31 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
5KO9
 
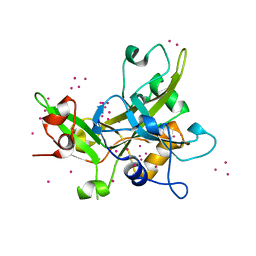 | | Crystal Structure of the SRAP Domain of Human HMCES Protein | | Descriptor: | Embryonic stem cell-specific 5-hydroxymethylcytosine-binding protein, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Li, Y, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-29 | | Release date: | 2016-08-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of HMCES interactions with abasic DNA and multivalent substrate recognition.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5W0F
 
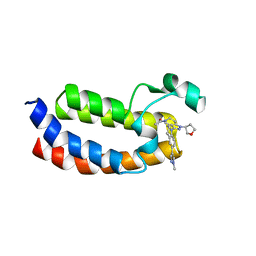 | | CREBBP Bromodomain in complex with Cpd3 ((S)-1-(3-(6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-1-(tetrahydrofuran-3-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | Descriptor: | 1-{3-[6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-1-[(3S)-oxolan-3-yl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, CREB-binding protein | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-30 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
5W0L
 
 | | CREBBP Bromodomain in complex with Cpd10 (1-(3-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-1-(tetrahydro-2H-pyran-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | Descriptor: | 1-{3-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-1-(oxan-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, CREB-binding protein | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-31 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
7F69
 
 | | Crystal structure of WIPI2b in complex with ATG16L1 | | Descriptor: | Isoform 2 of Autophagy-related protein 16-1, Isoform 2 of WD repeat domain phosphoinositide-interacting protein 2 | | Authors: | Gong, X.Y, Pan, L.F. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | ATG16L1 adopts a dual-binding site mode to interact with WIPI2b in autophagy.
Sci Adv, 9, 2023
|
|
4IXH
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Cryptosporidium parvum | | Descriptor: | (2S)-2-(naphthalen-1-yloxy)-N-[2-(pyridin-4-yl)-1,3-benzoxazol-5-yl]propanamide, 1,2-ETHANEDIOL, INOSINIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-25 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Optimization of Benzoxazole-Based Inhibitors of Cryptosporidium parvum Inosine 5'-Monophosphate Dehydrogenase.
J.Med.Chem., 56, 2013
|
|
4ZPE
 
 | |
6G3Z
 
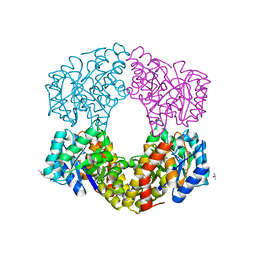 | | Sulfolobus sulfataricus 2-keto-3-deoxygluconate (KDG) aldolase complex with D-KDPG | | Descriptor: | 2-dehydro-3-deoxy-phosphogluconate/2-dehydro-3-deoxy-6-phosphogalactonate aldolase, 2-keto 3 deoxy 6 phospho gluconate, ISOPROPYL ALCOHOL | | Authors: | Crennell, S.J. | | Deposit date: | 2018-03-26 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Insights into the Substrate Specificity of Archaeal Entner-Doudoroff Aldolases: The Structures of Picrophilus torridus 2-Keto-3-deoxygluconate Aldolase and Sulfolobus solfataricus 2-Keto-3-deoxy-6-phosphogluconate Aldolase in Complex with 2-Keto-3-deoxy-6-phosphogluconate.
Biochemistry, 57, 2018
|
|
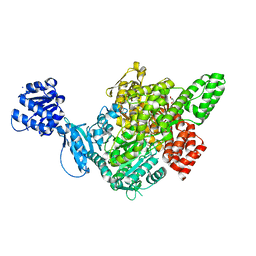
7C6O
 
 | |
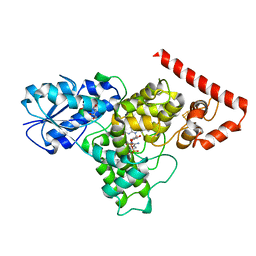
6DD6
 
 | |
5W0I
 
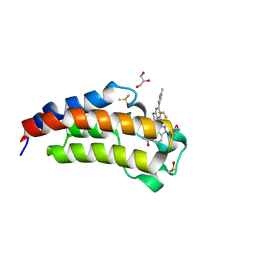 | | CREBBP Bromodomain in complex with Cpd8 (1-(3-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-1-(tetrahydrofuran-3-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | Descriptor: | 1-{3-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-1-[(3S)-oxolan-3-yl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, CREB-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-30 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
4JF6
 
 | | Structure of OXA-23 at pH 7.0 | | Descriptor: | Beta-lactamase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2013-02-27 | | Release date: | 2013-09-25 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Carbapenemase Activity of the OXA-23 beta-Lactamase from Acinetobacter baumannii.
Chem.Biol., 20, 2013
|
|
4ZPG
 
 | |
4ZHJ
 
 | |
4ZPF
 
 | |
6DKD
 
 | | Yeast Ddi2 Cyanamide Hydratase | | Descriptor: | DNA damage-inducible protein, SULFATE ION, ZINC ION | | Authors: | Moore, S.A, Xiao, W, Li, J. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Ddi2, a highly inducible detoxifying metalloenzyme fromSaccharomyces cerevisiae.
J.Biol.Chem., 294, 2019
|
|
4WV1
 
 | |
1D3Q
 
 | | CRYSTAL STRUCTURE OF HUMAN ALPHA THROMBIN IN COMPLEX WITH BENZO[B]THIOPHENE INHIBITOR 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[4-(2-PYRROLIDIN-1-YL-ETHOXY)-BENZYL]-2-4-(2-PYRROLIDIN-1-YL-ETHOXY)-PHENYL] -BENZO[B]THIOPHENE, ALPHA-THROMBIN, ... | | Authors: | Chirgadze, N.Y. | | Deposit date: | 1999-09-30 | | Release date: | 2000-10-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structures of human alpha-thrombin complexed with active site-directed diamino benzo[b]thiophene derivatives: a binding mode for a structurally novel class of inhibitors.
Protein Sci., 9, 2000
|
|
8OK4
 
 | | Variant Surface Glycoprotein VSG11wt-Oil | | Descriptor: | Variant surface glycoprotein, alpha-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zeelen, J.P, Stebbins, C.E, Aresta-Branco, F, Foti, K. | | Deposit date: | 2023-03-27 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | A structural classification of the variant surface glycoproteins of the African trypanosomey.
Plos Negl Trop Dis, 17, 2023
|
|
8ONH
 
 | | Variant Surface Glycoprotein VSG11wt-Oil | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Variant surface glycoprotein, alpha-D-glucopyranose, ... | | Authors: | Zeelen, J.P, Stebbins, C.E, Foti, K, Gkeka, A, Vlachou, E.P. | | Deposit date: | 2023-04-03 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A structural classification of the variant surface glycoproteins of the African trypanosomey.
Plos Negl Trop Dis, 17, 2023
|
|
6DD7
 
 | |
8OK5
 
 | | Variant Surface Glycoprotein VSG11 monomer with iodine | | Descriptor: | IODIDE ION, Variant surface glycoprotein, alpha-D-glucopyranose, ... | | Authors: | Zeelen, J.P, Stebbins, C.E, Aresta-Branco, F. | | Deposit date: | 2023-03-27 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | A structural classification of the variant surface glycoproteins of the African trypanosomey.
Plos Negl Trop Dis, 17, 2023
|
|
8OK6
 
 | | Variant Surface Glycoprotein VSG11 two monomers | | Descriptor: | SULFATE ION, Variant surface glycoprotein, alpha-D-glucopyranose, ... | | Authors: | Gkeka, A, Vlachou, E.P, Zeelen, J.P, Stebbins, C.E. | | Deposit date: | 2023-03-27 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A structural classification of the variant surface glycoproteins of the African trypanosomey.
Plos Negl Trop Dis, 17, 2023
|
|
8GI3
 
 | | Crystal structure of RhoA mutant L69P complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Chen, X, Qian, X, Chandravanshi, M, Lowy, D.R, Walters, K.J. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Ras-like GTPases mutants structure
To be published
|
|
