2HMC
 
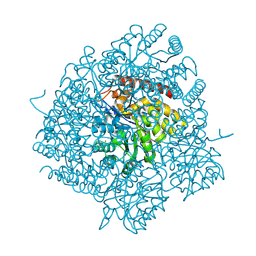 | | The Crystal Structure of Dihydrodipicolinate Synthase DapA from Agrobacterium tumefaciens | | Descriptor: | Dihydrodipicolinate synthase, MAGNESIUM ION | | Authors: | Kim, Y, Zhang, R, Xu, X, Zheng, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-11 | | Release date: | 2006-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Dihydrodipicolinate Synthase DapA from Agrobacterium tumefaciens
To be Published, 2006
|
|
2IJC
 
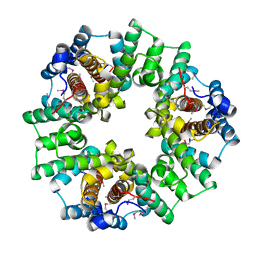 | | Structure of a Conserved Protein of Unknown Function PA0269 from Pseudomonas aeruginosa | | Descriptor: | Hypothetical protein | | Authors: | Binkowski, T.A, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-29 | | Release date: | 2006-12-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Crystal Structure of a Conserved Hypothetical Protein PA0269 from Pseudomonas aeruginosa
To be Published
|
|
2FRE
 
 | | The crystal structure of the oxidoreductase containing FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, NAD(P)H-flavin oxidoreductase | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-19 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the oxidoreductase containing FMN from Agrobacterium tumefaciens
To be Published
|
|
2I6H
 
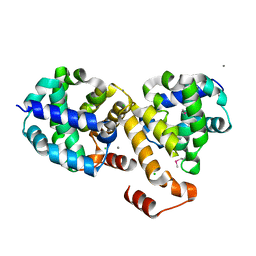 | | Structure of Protein of Unknown Function ATU0120 from Agrobacterium tumefaciens | | Descriptor: | CALCIUM ION, CHLORIDE ION, Hypothetical protein Atu0120 | | Authors: | Osipiuk, J, Xu, X, Gu, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-28 | | Release date: | 2006-09-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray crystal structure of hypothetical protein Atu0120 from Agrobacterium tumefaciens.
To be Published
|
|
2I10
 
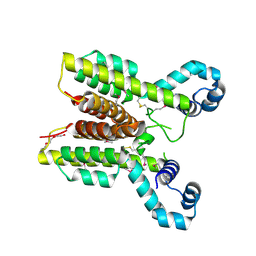 | | Putative TetR transcriptional regulator from Rhodococcus sp. RHA1 | | Descriptor: | P-NITROPHENOL, Putative TetR transcriptional regulator, TRIETHYLENE GLYCOL | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-11 | | Release date: | 2006-09-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray crystal structure of putative TetR transcriptional regulator from Rhodococcus sp. RHA1.
To be Published
|
|
2HYJ
 
 | | The crystal structure of a tetR-family transcriptional regulator from Streptomyces coelicolor | | Descriptor: | CALCIUM ION, Putative tetR-family transcriptional regulator, SULFATE ION | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-06 | | Release date: | 2006-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The crystal structure of a tetR-family transcriptional regulator from Streptomyces coelicolor
To be Published, 2006
|
|
2FSQ
 
 | | Crystal Structure of the Conserved Protein of Unknown Function ATU0111 from Agrobacterium tumefaciens str. C58 | | Descriptor: | ACETIC ACID, Atu0111 protein | | Authors: | Kim, Y, Joachimiak, A, Xu, X, Zheng, H, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the Conserved Hypothetical Protein Atu0111 from Agrobacterium tumefaciens str. C58
To be Published
|
|
2FDR
 
 | | Crystal Structure of Conserved Haloacid Dehalogenase-like Protein of Unknown Function ATU0790 from Agrobacterium tumefaciens str. C58 | | Descriptor: | MAGNESIUM ION, conserved hypothetical protein | | Authors: | Nocek, B, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-31 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of hydrolases/phosphatases-like fold protein
from Agrobacterium tumefaciens str. C58
To be Published
|
|
4PF1
 
 | | Crystal structure of aminopeptidase from marine sediment archaeon Thaumarchaeota archaeon | | Descriptor: | GLYCEROL, Peptidase S15/CocE/NonD, TRIETHYLENE GLYCOL | | Authors: | Michalska, K, Chhor, G, Fayman, K, Endres, M, Jedrzejczak, R, Babnigg, G, Steen, A, Lloyd, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-25 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New aminopeptidase from "microbial dark matter" archaeon.
FASEB J., 29, 2015
|
|
2GFQ
 
 | | Structure of Protein of Unknown Function PH0006 from Pyrococcus horikoshii | | Descriptor: | MAGNESIUM ION, SULFATE ION, UPF0204 protein PH0006 | | Authors: | Cuff, M.E, Skarina, T, Gorodichtchenskaia, E, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-22 | | Release date: | 2006-04-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of hypothetical protein ph0006 from Pyrococcus horikoshii
To be Published
|
|
2H8O
 
 | | The 1.6A crystal structure of the geranyltransferase from Agrobacterium tumefaciens | | Descriptor: | Geranyltranstransferase | | Authors: | Zhang, R, Xu, X, Gu, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-06-07 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6A crystal structure of the geranyltransferase from Agrobacterium tumefaciens
To be Published
|
|
1ZFJ
 
 | | INOSINE MONOPHOSPHATE DEHYDROGENASE (IMPDH; EC 1.1.1.205) FROM STREPTOCOCCUS PYOGENES | | Descriptor: | INOSINE MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID | | Authors: | Zhang, R, Evans, G, Rotella, F.J, Westbrook, E.M, Beno, D, Huberman, E, Joachimiak, A, Collart, F.R. | | Deposit date: | 1999-03-29 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characteristics and crystal structure of bacterial inosine-5'-monophosphate dehydrogenase.
Biochemistry, 38, 1999
|
|
3B4S
 
 | |
2FSR
 
 | | Crystal Structure of the Acetyltransferase from Agrobacterium tumefaciens str. C58 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, acetyltransferase | | Authors: | Kim, Y, Joachimiak, A, Xu, X, Gu, J, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of the Acetyltransferase from Agrobacterium tumefaciens str. C58
To be Published
|
|
2G39
 
 | | Crystal structure of coenzyme A transferase from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Acetyl-CoA hydrolase | | Authors: | Chang, C, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-02-17 | | Release date: | 2006-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of coenzyme A transferase from Pseudomonas aeruginosa
To be Published
|
|
3B49
 
 | |
3B85
 
 | |
3B4Q
 
 | |
3B79
 
 | |
2GYQ
 
 | | YcfI, a putative structural protein from Rhodopseudomonas palustris. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FE (III) ION, ... | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-05-09 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of YcfI protein, a putative structural protein from Rhodopseudomonas palustris.
To be Published
|
|
2GZ4
 
 | | 1.5 A Crystal Structure of a Protein of Unknown Function ATU1052 from Agrobacterium tumefaciens | | Descriptor: | Hypothetical protein Atu1052 | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-05-10 | | Release date: | 2006-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5A crystal structure of a hypothetical protein Atu1052 from Agrobacterium tumefaciens
To be Published
|
|
2GMY
 
 | | Crystal Structure of a Protein of Unknown Function ATU0492 from Agrobacterium tumefaciens, Putative Antioxidant Defence Protein AhpD | | Descriptor: | Hypothetical protein Atu0492 | | Authors: | Zhang, R, Xu, X, Gu, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of a hypothetical protein Atu0492 from Agrobacterium tumefaciens
To be Published
|
|
3BJN
 
 | | Crystal structure of C-terminal domain of putative transcriptional regulator from Vibrio cholerae, targeted domain 79-240 | | Descriptor: | CHLORIDE ION, Transcriptional regulator, putative | | Authors: | Chang, C, Volkart, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of C-terminal domain of putative transcriptional regulator from Vibrio cholerae.
To be Published
|
|
3BEE
 
 | | Crystal structure of putative YfrE protein from Vibrio parahaemolyticus | | Descriptor: | 1,2-ETHANEDIOL, Putative YfrE protein | | Authors: | Chang, C, Hatzos, C, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-16 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of putative YfrE protein from Vibrio parahaemolyticus.
To be Published
|
|
3BHG
 
 | | Crystal structure of adenylosuccinate lyase from Legionella pneumophila | | Descriptor: | Adenylosuccinate lyase, GLYCEROL, SULFATE ION | | Authors: | Chang, C, Li, H, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-28 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of adenylosuccinate lyase from Legionella pneumophila.
To be Published
|
|
