1ZOG
 
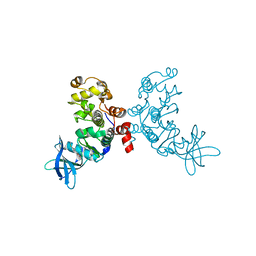 | | Crystal structure of protein kinase CK2 in complex with TBB-derivatives | | Descriptor: | 4,5,6,7-TETRABROMO-2-(METHYLSULFANYL)-1H-BENZIMIDAZOLE, PROTEIN KINASE CK2, alpha SUBUNIT | | Authors: | Battistutta, R, Mazzorana, M, Sarno, S, Kazimierczuk, Z, Zanotti, G, Pinna, L.A. | | Deposit date: | 2005-05-13 | | Release date: | 2005-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inspecting the structure-activity relationship of protein kinase CK2 inhibitors derived from tetrabromo-benzimidazole.
Chem.Biol., 12, 2005
|
|
3HPE
 
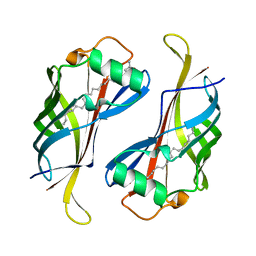 | |
1ZOF
 
 | | Crystal structure of alkyl hydroperoxide-reductase (AhpC) from Helicobacter Pylori | | Descriptor: | alkyl hydroperoxide-reductase | | Authors: | Papinutto, E, Windle, H.J, Cendron, L, Battistutta, R, Kelleher, D, Zanotti, G. | | Deposit date: | 2005-05-13 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of alkyl hydroperoxide-reductase (AhpC) from Helicobacter pylori.
Biochim.Biophys.Acta, 1753, 2005
|
|
3CL8
 
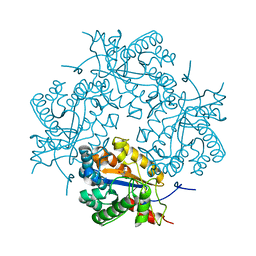 | | Crystal structure of Puue Allantoinase complexed with ACA | | Descriptor: | 5-amino-1H-imidazole-4-carboxamide, Puue Allantoinase | | Authors: | Ramazzina, I, Cendron, L, Folli, C, Berni, R, Monteverdi, D, Zanotti, G, Percudani, R. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Logical identification of an allantoinase analog (puuE) recruited from polysaccharide deacetylases
J.Biol.Chem., 283, 2008
|
|
3CL7
 
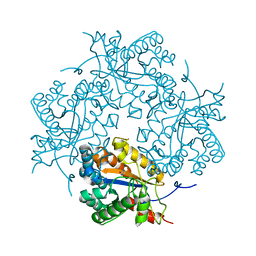 | | Crystal structure of Puue Allantoinase in complex with Hydantoin | | Descriptor: | Puue Allantoinase, imidazolidine-2,4-dione | | Authors: | Ramazzina, I, Cendron, L, Folli, C, Berni, R, Monteverdi, D, Zanotti, G, Percudani, R. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Logical identification of an allantoinase analog (puuE) recruited from polysaccharide deacetylases
J.Biol.Chem., 283, 2008
|
|
3CL6
 
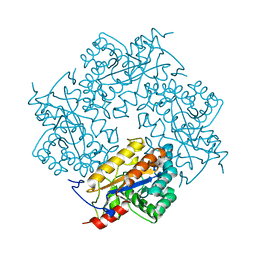 | | Crystal structure of Puue Allantoinase | | Descriptor: | Puue allantoinase | | Authors: | Ramazzina, I, Cendron, L, Folli, C, Berni, R, Monteverdi, D, Zanotti, G, Percudani, R. | | Deposit date: | 2008-03-18 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Logical identification of an allantoinase analog (puuE) recruited from polysaccharide deacetylases
J.Biol.Chem., 283, 2008
|
|
2ALG
 
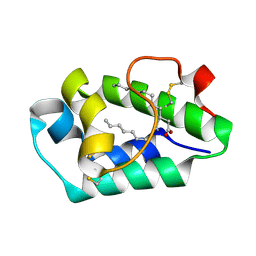 | | Crystal structure of peach Pru p3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens | | Descriptor: | HEPTANE, HEXAETHYLENE GLYCOL, LAURIC ACID, ... | | Authors: | Pasquato, N, Berni, R, Folli, C, Folloni, S, Cianci, M, Pantano, S, Helliwell, J, Zanotti, G. | | Deposit date: | 2005-08-05 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Peach Pru p 3, the Prototypic Member of the Family of Plant Non-specific Lipid Transfer Protein Pan-allergens
J.Mol.Biol., 356, 2006
|
|
3QQ5
 
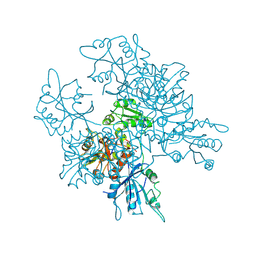 | | Crystal structure of the [FeFe]-hydrogenase maturation protein HydF | | Descriptor: | Small GTP-binding protein | | Authors: | Cendron, L, Berto, P, D'Adamo, S, Vallese, F, Govoni, C, Posewitz, M.C, Giacometti, G.M, Costantini, P, Zanotti, G. | | Deposit date: | 2011-02-15 | | Release date: | 2011-11-16 | | Last modified: | 2012-01-11 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal Structure of HydF Scaffold Protein Provides Insights into [FeFe]-Hydrogenase Maturation.
J.Biol.Chem., 286, 2011
|
|
3QBU
 
 | | Crystal structure of putative peptidoglycan deactelyase (HP0310) from Helicobacter pylori | | Descriptor: | Putative uncharacterized protein, ZINC ION | | Authors: | Shaik, M.M, Cendron, L, Percudani, R, Zanotti, G. | | Deposit date: | 2011-01-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5701 Å) | | Cite: | The Structure of Helicobacter pylori HP0310 Reveals an Atypical Peptidoglycan Deacetylase.
Plos One, 6, 2011
|
|
3TLM
 
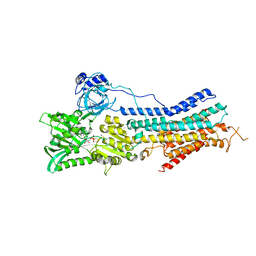 | | Crystal Structure of Endoplasmic Reticulum Ca2+-ATPase (SERCA) From Bovine Muscle | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Sacchetto, R, Bertipaglia, I, Giannetti, S, Cendron, L, Mascarello, F, Damiani, E, Carafoli, E, Zanotti, G. | | Deposit date: | 2011-08-30 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of sarcoplasmic reticulum Ca(2+)-ATPase (SERCA) from bovine muscle.
J.Struct.Biol., 178, 2012
|
|
3SXP
 
 | |
6Z05
 
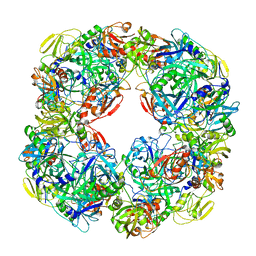 | |
1ORB
 
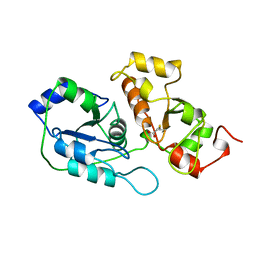 | | ACTIVE SITE STRUCTURAL FEATURES FOR CHEMICALLY MODIFIED FORMS OF RHODANESE | | Descriptor: | ACETATE ION, CARBOXYMETHYLATED RHODANESE | | Authors: | Gliubich, F, Gazerro, M, Zanotti, G, Delbono, S, Berni, R. | | Deposit date: | 1995-07-24 | | Release date: | 1995-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active site structural features for chemically modified forms of rhodanese.
J.Biol.Chem., 271, 1996
|
|
4INO
 
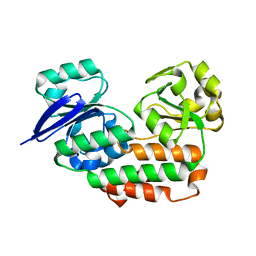 | |
4INN
 
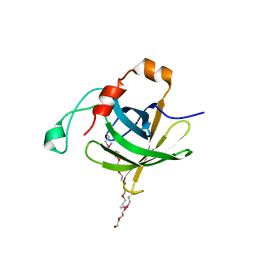 | | Protein HP1028 from the human pathogen Helicobacter pylori belongs to the lipocalin family | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, HP1028 | | Authors: | Barison, N, Cendron, L, Loconte, V, Zanotti, G. | | Deposit date: | 2013-01-05 | | Release date: | 2013-07-31 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein HP1028 from the human pathogen Helicobacter pylori belongs to the lipocalin family.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4INP
 
 | | The crystal structure of Helicobacter pylori Ceue (HP1561) with Ni(II) bound | | Descriptor: | ACETATE ION, Iron (III) ABC transporter, periplasmic iron-binding protein, ... | | Authors: | Shaik, M.M, Cendron, L, Zanotti, G. | | Deposit date: | 2013-01-05 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Helicobacter pylori periplasmic receptor CeuE (HP1561) modulates its nickel affinity via organic metallophores.
Mol.Microbiol., 91, 2014
|
|
2ORA
 
 | | RHODANESE (THIOSULFATE: CYANIDE SULFURTRANSFERASE) | | Descriptor: | OXIDIZED RHODANESE | | Authors: | Gliubich, F, Gazerro, M, Zanotti, G, Delbono, S, Berni, R. | | Deposit date: | 1996-02-22 | | Release date: | 1996-08-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Active site structural features for chemically modified forms of rhodanese.
J.Biol.Chem., 271, 1996
|
|
4MNT
 
 | | Crystal structure of human DJ-1 in complex with Cu | | Descriptor: | COPPER (II) ION, Protein DJ-1 | | Authors: | Cendron, L, Girotto, S, Bisaglia, M, Tessari, I, Mammi, S, Zanotti, G, Bubacco, L. | | Deposit date: | 2013-09-11 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.584 Å) | | Cite: | DJ-1 Is a Copper Chaperone Acting on SOD1 Activation.
J.Biol.Chem., 289, 2014
|
|
4KZS
 
 | | Crystal structure of the secreted protein HP1454 from the human pathogen Helicobacter pylori | | Descriptor: | LPP20 lipofamily protein, PENTAETHYLENE GLYCOL | | Authors: | Quarantini, S, Cendron, L, Fischer, W, Zanotti, G. | | Deposit date: | 2013-05-30 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the secreted protein HP1454 from the human pathogen Helicobacter pylori.
Proteins, 82, 2014
|
|
3ISL
 
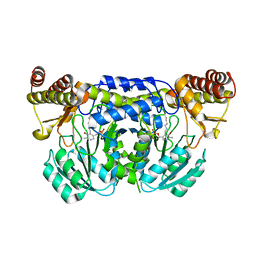 | | Crystal structure of ureidoglycine-glyoxylate aminotransferase (pucG) from Bacillus subtilis | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Purine catabolism protein pucG | | Authors: | Costa, R, Cendron, L, Ramazzina, I, Berni, R, Peracchi, A, Percudani, R, Zanotti, G. | | Deposit date: | 2009-08-26 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Amino acids from purines in GUT bacteria
To be Published
|
|
1KT4
 
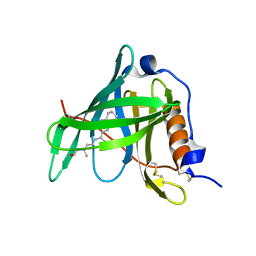 | | Crystal structure of bovine holo-RBP at pH 3.0 | | Descriptor: | RETINOL, plasma retinol-binding protein | | Authors: | Calderone, V, Berni, R, Zanotti, G. | | Deposit date: | 2002-01-15 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.461 Å) | | Cite: | High-resolution Structures of Retinol-binding Protein in
Complex with Retinol: pH-induced Protein Structural
Changes in the Crystal State
J.Mol.Biol., 329, 2003
|
|
1KT3
 
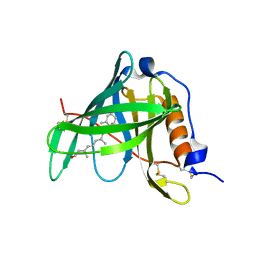 | | Crystal structure of bovine holo-RBP at pH 2.0 | | Descriptor: | Plasma retinol-binding protein, RETINOL | | Authors: | Calderone, V, Berni, R, Zanotti, G. | | Deposit date: | 2002-01-15 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution Structures of
Retinol-binding Protein in
Complex with Retinol: pH-induced Protein Structural
Changes in the Crystal State
J.Mol.Biol., 329, 2003
|
|
1KT7
 
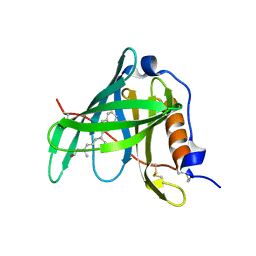 | | Crystal structure of bovine holo-RBP at pH 7.0 | | Descriptor: | Plasma retinol-binding protein, RETINOL | | Authors: | Calderone, V, Berni, R, Zanotti, G. | | Deposit date: | 2002-01-15 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.274 Å) | | Cite: | High-resolution Structures of Retinol-binding Protein in
Complex with Retinol: pH-induced Protein Structural
Changes in the Crystal State
J.Mol.Biol., 329, 2003
|
|
1KQX
 
 | | Crystal structure of apo-CRBP from zebrafish | | Descriptor: | Cellular retinol-binding protein | | Authors: | Calderone, V, Folli, C, Marchesani, A, Berni, R, Zanotti, G. | | Deposit date: | 2002-01-08 | | Release date: | 2002-08-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and structural analysis of a zebrafish apo and holo cellular retinol-binding protein.
J.Mol.Biol., 321, 2002
|
|
1KQW
 
 | | Crystal structure of holo-CRBP from zebrafish | | Descriptor: | Cellular retinol-binding protein, RETINOL | | Authors: | Calderone, V, Folli, C, Marchesani, A, Berni, R, Zanotti, G. | | Deposit date: | 2002-01-08 | | Release date: | 2002-08-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Identification and structural analysis of a zebrafish apo and holo cellular retinol-binding protein.
J.Mol.Biol., 321, 2002
|
|
