3BUQ
 
 | |
3CZN
 
 | | Golgi alpha-mannosidase II (D204A nucleophile mutant) in complex with GnMan5Gn | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Alpha-mannosidase 2, ... | | Authors: | Shah, N, Rose, D.R. | | Deposit date: | 2008-04-29 | | Release date: | 2008-06-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Golgi alpha-mannosidase II cleaves two sugars sequentially in the same catalytic site.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3D4Z
 
 | | GOLGI MANNOSIDASE II complex with gluco-imidazole | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Alpha-mannosidase 2, GLUCOIMIDAZOLE, ... | | Authors: | Kuntz, D.A, Tarling, C.A, Withers, S.G, Rose, D.R. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural analysis of Golgi alpha-mannosidase II inhibitors identified from a focused glycosidase inhibitor screen.
Biochemistry, 47, 2008
|
|
3D51
 
 | | GOLGI MANNOSIDASE II complex with gluco-hydroxyiminolactam | | Descriptor: | (2S,3S,4R,5R)-6-(HYDROXYAMINO)-2-(HYDROXYMETHYL)-2,3,4,5-TETRAHYDROPYRIDINE-3,4,5-TRIOL, (4R)-2-METHYLPENTANE-2,4-DIOL, Alpha-mannosidase 2, ... | | Authors: | Kuntz, D.A, Tarling, C.A, Withers, S.G, Rose, D.R. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural analysis of Golgi alpha-mannosidase II inhibitors identified from a focused glycosidase inhibitor screen.
Biochemistry, 47, 2008
|
|
3CUI
 
 | |
3CV5
 
 | | GOLGI MANNOSIDASE II D204A catalytic nucleophile mutant complex with 3alpha,6alpha-mannopentaose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-mannosidase 2, ... | | Authors: | Shah, N, Kuntz, D.A, Rose, D.R. | | Deposit date: | 2008-04-17 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Golgi alpha-mannosidase II cleaves two sugars sequentially in the same catalytic site.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3CZS
 
 | | Golgi alpha-mannosidase II (D204A nucleophile mutant) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-mannosidase 2, ... | | Authors: | Shah, N, Rose, D.R. | | Deposit date: | 2008-04-29 | | Release date: | 2008-06-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Golgi alpha-mannosidase II cleaves two sugars sequentially in the same catalytic site.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3CUH
 
 | |
3CUG
 
 | |
3D52
 
 | | GOLGI MANNOSIDASE II complex with an N-aryl carbamate derivative of gluco-hydroxyiminolactam | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Alpha-mannosidase 2, ZINC ION, ... | | Authors: | Kuntz, D.A, Tarling, C.A, Withers, S.G, Rose, D.R. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analysis of Golgi alpha-mannosidase II inhibitors identified from a focused glycosidase inhibitor screen.
Biochemistry, 47, 2008
|
|
4MCK
 
 | |
2QMJ
 
 | | Crystral Structure of the N-terminal Subunit of Human Maltase-Glucoamylase in Complex with Acarbose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2007-07-16 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human intestinal maltase-glucoamylase: crystal structure of the N-terminal catalytic subunit and basis of inhibition and substrate specificity
J.Mol.Biol., 375, 2008
|
|
2QLY
 
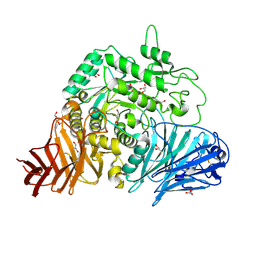 | | Crystral Structure of the N-terminal Subunit of Human Maltase-Glucoamylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2007-07-13 | | Release date: | 2008-01-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human intestinal maltase-glucoamylase: crystal structure of the N-terminal catalytic subunit and basis of inhibition and substrate specificity
J.Mol.Biol., 375, 2008
|
|
1MFC
 
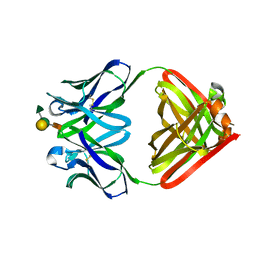 | |
1MFB
 
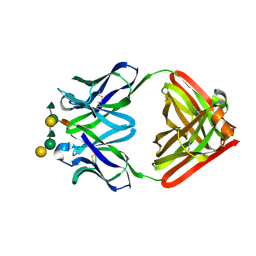 | |
8FAB
 
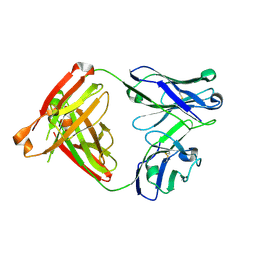 | |
1MFE
 
 | |
1MFA
 
 | |
1GUI
 
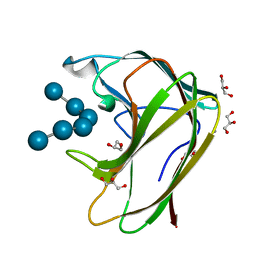 | | CBM4 structure and function | | Descriptor: | CALCIUM ION, GLYCEROL, LAMINARINASE 16A, ... | | Authors: | Nurizzo, D, Notenboom, V, Davies, G.J. | | Deposit date: | 2002-01-27 | | Release date: | 2002-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential Oligosaccharide Recognition by Evolutionarily-Related Beta-1,4 and Beta-1,3 Glucan-Binding Modules
J.Mol.Biol., 319, 2002
|
|
1GU3
 
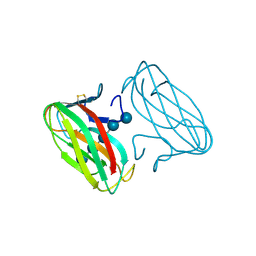 | | CBM4 structure and function | | Descriptor: | ENDOGLUCANASE C, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Nurizzo, D, Notenboom, V, Davies, G.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Differential Oligosaccharide Recognition by Evolutionarily-Related Beta-1,4 and Beta-1,3 Glucan-Binding Modules
J.Mol.Biol., 319, 2002
|
|
1MFD
 
 | |
1XNB
 
 | |
1THE
 
 | |
1IGJ
 
 | |
1IGI
 
 | |
