5ULY
 
 | |
2DWU
 
 | | Crystal Structure of Glutamate Racemase Isoform RacE1 from Bacillus anthracis | | Descriptor: | D-GLUTAMIC ACID, GLYCEROL, Glutamate racemase, ... | | Authors: | Mehboob, S, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2006-08-17 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Analysis of Two Glutamate Racemase Isozymes from Bacillus anthracis and Implications for Inhibitor Design
J.Mol.Biol., 371, 2007
|
|
3GR8
 
 | | Structure of OYE from Geobacillus kaustophilus, orthorhombic crystal form | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase, ... | | Authors: | Uhl, M.K, Gruber, K. | | Deposit date: | 2009-03-25 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Old Yellow Enzyme-Catalyzed Dehydrogenation of Saturated Ketones
ADV.SYNTH.CATAL., 353, 2011
|
|
6MER
 
 | | PcdhgB3 EC1-4 in 50 mM HEPES | | Descriptor: | CALCIUM ION, Protocadherin gamma-B3 | | Authors: | Nicoludis, J.M, Gaudet, R. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Interaction specificity of clustered protocadherins inferred from sequence covariation and structural analysis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6MEQ
 
 | | PcdhgB3 EC1-4 in 50 mM HEPES | | Descriptor: | CALCIUM ION, Protocadherin gamma-B3 | | Authors: | Nicoludis, J.M, Gaudet, R. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Interaction specificity of clustered protocadherins inferred from sequence covariation and structural analysis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6YYK
 
 | | Crystal Structure of 1,5-dimethylindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 1,5-dimethyl-3~{H}-indol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To Be Published
|
|
6YYF
 
 | | Crystal Structure of 5-chloroindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 5-chloranyl-1,3-dihydroindol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To Be Published
|
|
6YYG
 
 | | Crystal Structure of 5-(trifluoromethoxy)indoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 5-(trifluoromethyloxy)-1,3-dihydroindol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To Be Published
|
|
6TT2
 
 | |
6TSE
 
 | | Crystal Structure of 1-methylindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C | | Descriptor: | 1-methylindole-2,3-dione, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2019-12-20 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal Structure of 1-methylindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C
To Be Published
|
|
6TUH
 
 | | PH domain of Bruton's tyrosine kinase bound to compound 1 | | Descriptor: | 4,5,6,7-tetrahydro-1-benzofuran-3-carboxylic acid, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of 1-methylindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C
To Be Published
|
|
6TVN
 
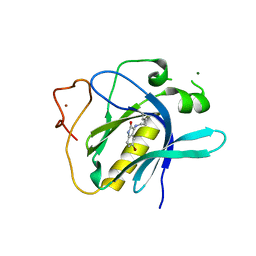 | | Crystal Structure of 5-bromoindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase | | Descriptor: | 5-bromanyl-1,3-dihydroindol-2-one, MAGNESIUM ION, Tyrosine-protein kinase BTK, ... | | Authors: | Brear, P, Wagstaff, J, Hyvonen, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of 1-methylindoline-2,3-dione covalently bound to the PH domain of Bruton's tyrosine kinase mutant R28C
To Be Published
|
|
6CWA
 
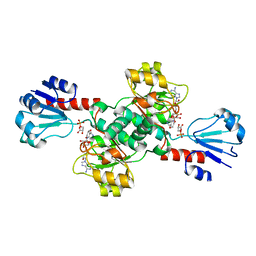 | |
6W1J
 
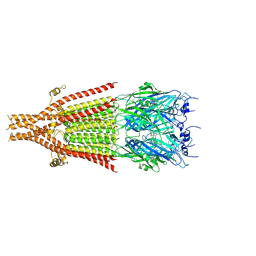 | | Cryo-EM structure of 5HT3A receptor in presence of Alosetron | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(4-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, ... | | Authors: | Basak, S, Chakrapani, S. | | Deposit date: | 2020-03-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | High-resolution structures of multiple 5-HT 3A R-setron complexes reveal a novel mechanism of competitive inhibition.
Elife, 9, 2020
|
|
5DKA
 
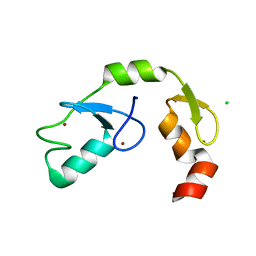 | | A C2HC zinc finger is essential for the activity of the RING ubiquitin ligase RNF125 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF125, MAGNESIUM ION, ... | | Authors: | Boer, D.R, Coll, M, Bijlmakers, M.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A C2HC zinc finger is essential for the RING-E2 interaction of the ubiquitin ligase RNF125.
Sci Rep, 6, 2016
|
|
6Q7P
 
 | | Crystal structure of OE1.2 | | Descriptor: | 1,2-ETHANEDIOL, 1-PHENYLETHANONE, MAGNESIUM ION, ... | | Authors: | Levy, C.W. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Design and evolution of an enzyme with a non-canonical organocatalytic mechanism.
Nature, 570, 2019
|
|
6Q7R
 
 | |
6Q7O
 
 | | Crystal structure of OE1 | | Descriptor: | CALCIUM ION, OE1 | | Authors: | Levy, C.W. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and evolution of an enzyme with a non-canonical organocatalytic mechanism.
Nature, 570, 2019
|
|
6Q7N
 
 | |
6Q7Q
 
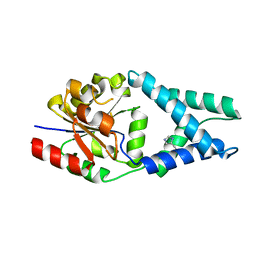 | | Crystal structure of OE1.3 | | Descriptor: | OE1.3 | | Authors: | Levy, C.W. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and evolution of an enzyme with a non-canonical organocatalytic mechanism.
Nature, 570, 2019
|
|
2BZT
 
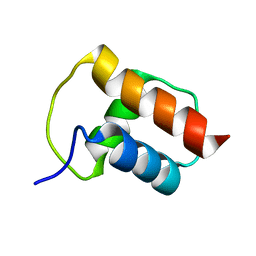 | | NMR structure of the bacterial protein YFHJ from E. coli | | Descriptor: | PROTEIN ISCX | | Authors: | Pastore, C, Kelly, G, Adinolfi, S, Mc Cormick, J.E, Pastore, A. | | Deposit date: | 2005-08-22 | | Release date: | 2006-12-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | YfhJ, a molecular adaptor in iron-sulfur cluster formation or a frataxin-like protein?
Structure, 14, 2006
|
|
7O7B
 
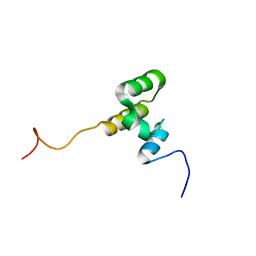 | |
9EQF
 
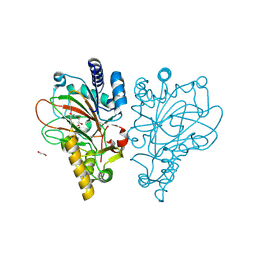 | | Crystal structure of the L-arginine hydroxylase VioC MeHis316, bound to Fe(II), L-arginine, and succinate | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, Alpha-ketoglutarate-dependent L-arginine hydroxylase, ... | | Authors: | Hardy, F.J. | | Deposit date: | 2024-03-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing Ferryl Reactivity in a Nonheme Iron Oxygenase Using an Expanded Genetic Code.
Acs Catalysis, 14, 2024
|
|
9FWX
 
 | | Crystal structure of BRD4 BD1 with MEN1404BS. | | Descriptor: | 3-methyl-~{N}-(2-phenylethyl)-[1,2,4]triazolo[4,3-b]pyridazin-6-amine, Bromodomain-containing protein 4 | | Authors: | Bader, G, Kessler, D. | | Deposit date: | 2024-07-01 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Probing Protein-Ligand Methyl-pi Interaction Geometries through Chemical Shift Measurements of Selectively Labeled Methyl Groups.
J.Med.Chem., 67, 2024
|
|
9FXP
 
 | | Crystal structure of BRD4 BD1 with DI00626383. | | Descriptor: | 4-methoxy-1,2-benzoxazol-3-amine, Bromodomain-containing protein 4 | | Authors: | Bader, G, Reinert, D. | | Deposit date: | 2024-07-02 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Probing Protein-Ligand Methyl-pi Interaction Geometries through Chemical Shift Measurements of Selectively Labeled Methyl Groups.
J.Med.Chem., 67, 2024
|
|
