1IIK
 
 | | CRYSTAL STRUCTURE OF THE TRANSTHYRETIN MUTANT TTR Y114C-DATA COLLECTED AT CRYO TEMPERATURE | | Descriptor: | BETA-MERCAPTOETHANOL, TRANSTHYRETIN | | Authors: | Eneqvist, T, Olofsson, A, Ando, Y, Lundgren, E, Sauer-Eriksson, A.E. | | Deposit date: | 2001-04-23 | | Release date: | 2002-11-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disulfide-Bond Formation in the Transthyretin Mutant Y114C Prevents Amyloid Fibril Formation in Vivo and in Vitro
Biochemistry, 41, 2002
|
|
1FCC
 
 | | CRYSTAL STRUCTURE OF THE C2 FRAGMENT OF STREPTOCOCCAL PROTEIN G IN COMPLEX WITH THE FC DOMAIN OF HUMAN IGG | | Descriptor: | IGG1 MO61 FC, STREPTOCOCCAL PROTEIN G (C2 FRAGMENT) | | Authors: | Sauer-Eriksson, A.E, Kleywegt, G.J, Uhlen, M, Jones, T.A. | | Deposit date: | 1995-01-17 | | Release date: | 1995-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the C2 fragment of streptococcal protein G in complex with the Fc domain of human IgG.
Structure, 3, 1995
|
|
1G1O
 
 | | CRYSTAL STRUCTURE OF THE HIGHLY AMYLOIDOGENIC TRANSTHYRETIN MUTANT TTR G53S/E54D/L55S | | Descriptor: | TRANSTHYRETIN | | Authors: | Eneqvist, T, Andersson, K, Olofsson, A, Lundgren, E, Sauer-Eriksson, A.E. | | Deposit date: | 2000-10-13 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The beta-slip: a novel concept in transthyretin amyloidosis.
Mol.Cell, 6, 2000
|
|
1SOK
 
 | | Crystal structure of the transthyretin mutant A108Y/L110E solved in space group p21212 | | Descriptor: | Transthyretin | | Authors: | Hornberg, A, Olofsson, A, Eneqvist, T, Lundgren, E, Sauer-Eriksson, A.E. | | Deposit date: | 2004-03-15 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The beta-strand D of transthyretin trapped in two discrete conformations
Biochim.Biophys.Acta, 1700, 2004
|
|
1SN5
 
 | | Crystal Structure of Sea Bream Transthyretin in complex with Triiodothyronine at 1.90A Resolution | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, SULFATE ION, transthyretin | | Authors: | Eneqvist, T, Lundberg, E, Karlsson, A, Huang, S, Cantos, C.R, Power, D.M, Sauer-Eriksson, A.E. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution crystal structures of piscine transthyretin reveal different binding modes for triiodothyronine and thyroxine.
J.Biol.Chem., 279, 2004
|
|
1SN2
 
 | | Crystal Structure of Sea Bream Transthyretin at 1.90A Resolution | | Descriptor: | SULFATE ION, transthyretin | | Authors: | Eneqvist, T, Lundberg, E, Karlsson, A, Huang, S, Cantos, C.R, Power, D.M, Sauer-Eriksson, A.E. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High resolution crystal structures of piscine transthyretin reveal different binding modes for triiodothyronine and thyroxine.
J.Biol.Chem., 279, 2004
|
|
1SN0
 
 | | Crystal Structure Of Sea Bream Transthyretin in complex with thyroxine At 1.9A Resolution | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, SULFATE ION, transthyretin | | Authors: | Eneqvist, T, Lundberg, E, Karlsson, A, Huang, S, Santos, C.R, Power, D.M, Sauer-Eriksson, A.E. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution crystal structures of piscine transthyretin reveal different binding modes for triiodothyronine and thyroxine.
J.Biol.Chem., 279, 2004
|
|
1SOQ
 
 | | Crystal structure of the transthyretin mutant A108Y/L110E solved in space group C2 | | Descriptor: | Transthyretin | | Authors: | Hornberg, A, Olofsson, A, Eneqvist, T, Lundgren, E, Sauer-Eriksson, A.E. | | Deposit date: | 2004-03-15 | | Release date: | 2004-07-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The beta-strand D of transthyretin trapped in two discrete conformations
Biochim.Biophys.Acta, 1700, 2004
|
|
4L9D
 
 | | Crystal structure of the PKD1 domain from Vibrio cholerae metalloprotease PrtV | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Edwin, A, Stier, G, Sauer-Eriksson, A.E. | | Deposit date: | 2013-06-18 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Calcium binding by the PKD1 domain regulates interdomain flexibility in Vibrio cholerae metalloprotease PrtV.
FEBS Open Bio, 3, 2013
|
|
1Z43
 
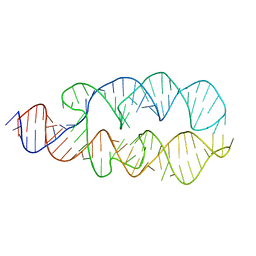 | |
1LNG
 
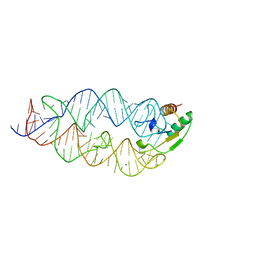 | |
1LGD
 
 | | Crystal Structure Analysis of HCA II Mutant T199P in Complex with Bicarbonate | | Descriptor: | BICARBONATE ION, Carbonic anhydrase II, ZINC ION | | Authors: | Huang, S, Sjoblom, B, Sauer-Eriksson, A.E, Jonsson, B.-H. | | Deposit date: | 2002-04-15 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Organization of an efficient carbonic anhydrase: implications for the mechanism based on structure-function studies of a T199P/C206S mutant.
Biochemistry, 41, 2002
|
|
1LG6
 
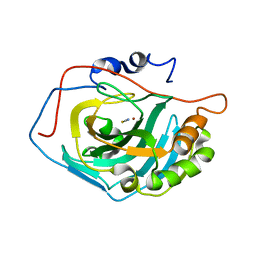 | | Crystal Structure Analysis of HCA II Mutant T199P in Complex with Thiocyanate | | Descriptor: | Carbonic anhydrase II, THIOCYANATE ION, ZINC ION | | Authors: | Huang, S, Sjoblom, B, Sauer-Eriksson, A.E, Jonsson, B.-H. | | Deposit date: | 2002-04-15 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Organization of an efficient carbonic anhydrase: implications for the mechanism based on structure-function studies of a T199P/C206S mutant.
Biochemistry, 41, 2002
|
|
1LG5
 
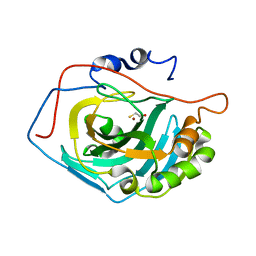 | | Crystal Structure Analysis of the HCA II Mutant T199P in complex with beta-mercaptoethanol | | Descriptor: | BETA-MERCAPTOETHANOL, Carbonic anhydrase II, ZINC ION | | Authors: | Huang, S, Sjoblom, B, Sauer-Eriksson, A.E, Jonsson, B.-H. | | Deposit date: | 2002-04-15 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Organization of an efficient carbonic anhydrase: implications for the mechanism based on structure-function studies of a T199P/C206S mutant.
Biochemistry, 41, 2002
|
|
1ZU4
 
 | |
1ZD6
 
 | | Crystal structure of human transthyretin with bound chloride | | Descriptor: | CHLORIDE ION, Transthyretin | | Authors: | Hornberg, A, Hultdin, U.W, Olofsson, A, Sauer-Eriksson, A.E. | | Deposit date: | 2005-04-14 | | Release date: | 2005-07-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The effect of iodide and chloride on transthyretin structure and stability
Biochemistry, 44, 2005
|
|
1ZU5
 
 | |
1ZCR
 
 | | Crystal structure of human Transthyretin with bound iodide | | Descriptor: | IODIDE ION, Transthyretin | | Authors: | Hornberg, A, Hultdin, U.W, Olofsson, A, Sauer-Eriksson, A.E. | | Deposit date: | 2005-04-13 | | Release date: | 2005-07-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The effect of iodide and chloride on transthyretin structure and stability
Biochemistry, 44, 2005
|
|
5CAC
 
 | | REFINED STRUCTURE OF HUMAN CARBONIC ANHYDRASE II AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CARBONIC ANHYDRASE FORM C, SULFITE ION, ZINC ION | | Authors: | Lindahl, M, Habash, D, Harrop, S, Helliwell, D.R, Liljas, A. | | Deposit date: | 1991-09-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined structure of human carbonic anhydrase II at 2.0 A resolution.
Proteins, 4, 1988
|
|
4CAC
 
 | | REFINED STRUCTURE OF HUMAN CARBONIC ANHYDRASE II AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CARBONIC ANHYDRASE FORM C, ZINC ION | | Authors: | Lindahl, M, Habash, D, Harrop, S, Helliwell, D.R, Liljas, A. | | Deposit date: | 1991-09-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined structure of human carbonic anhydrase II at 2.0 A resolution.
Proteins, 4, 1988
|
|
4LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
3NDB
 
 | | Crystal structure of a signal sequence bound to the signal recognition particle | | Descriptor: | PHOSPHATE ION, SRP RNA, Signal recognition 54 kDa protein, ... | | Authors: | Hainzl, T, Huang, S, Sauer-Eriksson, E. | | Deposit date: | 2010-06-07 | | Release date: | 2011-02-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of signal-sequence recognition by the signal recognition particle.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
7LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
5LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
