1Q5W
 
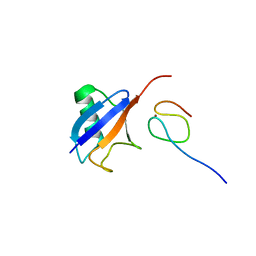 | | Ubiquitin Recognition by Npl4 Zinc-Fingers | | Descriptor: | Ubiquitin, ZINC ION, homolog of yeast nuclear protein localization 4 | | Authors: | Alam, S.L, Sun, J, Payne, M, Welch, B.D, Blake, B.K, Davis, D.R, Meyer, H.H, Emr, S.D, Sundquist, W.I. | | Deposit date: | 2003-08-11 | | Release date: | 2004-03-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin interactions of NZF zinc fingers.
Embo J., 23, 2004
|
|
2C4Y
 
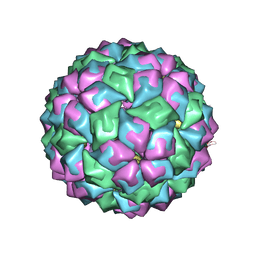 | | MS2-RNA HAIRPIN (2THIOURACIL-5) COMPLEX | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *SUR*AP*CP*CP*CP*AP*UP*GP*U)-3', Capsid protein | | Authors: | Grahn, E, Moss, T, Helgstrand, C, Fridborg, K, Sundaram, M, Tars, K, Lago, H, Stonehouse, N.J, Davis, D.R, Stockley, P.G, Liljas, L. | | Deposit date: | 2005-10-25 | | Release date: | 2006-01-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis of pyrimidine specificity in the MS2 RNA hairpin-coat-protein complex.
Rna, 7, 2001
|
|
2C50
 
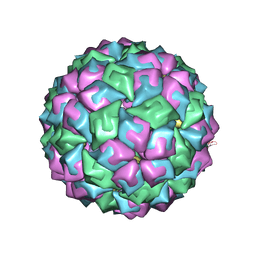 | | MS2-RNA HAIRPIN (A -5) COMPLEX | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *AP*AP*CP*CP*CP*AP*UP*GP*U)-3', COAT PROTEIN | | Authors: | Grahn, E, Moss, T, Helgstrand, C, Fridborg, K, Sundaram, M, Tars, K, Lago, H, Stonehouse, N.J, Davis, D.R, Stockley, P.G, Liljas, L. | | Deposit date: | 2005-10-25 | | Release date: | 2006-01-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of pyrimidine specificity in the MS2 RNA hairpin-coat-protein complex.
Rna, 7, 2001
|
|
2C4Z
 
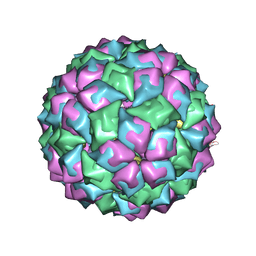 | | MS2-RNA HAIRPIN (2SU -5-6) COMPLEX | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*SUR *SUR*AP*CP*CP*CP*AP*UP*GP*U)-3', COAT PROTEIN | | Authors: | Grahn, E, Moss, T, Helgstrand, C, Fridborg, K, Sundaram, M, Tars, K, Lago, H, Stonehouse, N.J, Davis, D.R, Stockley, P.G, Liljas, L. | | Deposit date: | 2005-10-25 | | Release date: | 2006-01-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of pyrimidine specificity in the MS2 RNA hairpin-coat-protein complex.
Rna, 7, 2001
|
|
2C4Q
 
 | | MS2-RNA HAIRPIN (2ONE -5) COMPLEX | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *PYO*AP*CP*CP*CP*AP*UP*GP*U)-3', COAT PROTEIN | | Authors: | Grahn, E, Moss, T, Helgstrand, C, Fridborg, K, Sundaram, M, Tars, K, Lago, H, Stonehouse, N.J, Davis, D.R, Stockley, P.G, Liljas, L. | | Deposit date: | 2005-10-21 | | Release date: | 2005-11-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis of pyrimidine specificity in the MS2 RNA hairpin-coat-protein complex.
Rna, 7, 2001
|
|
2C51
 
 | | MS2-RNA HAIRPIN (G -5) COMPLEX | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *GP*AP*CP*CP*CP*AP*UP*GP*U)-3', COAT PROTEIN | | Authors: | Grahn, E, Moss, T, Helgstrand, C, Fridborg, K, Sundaram, M, Tars, K, Lago, H, Stonehouse, N.J, Davis, D.R, Stockley, P.G, Liljas, L. | | Deposit date: | 2005-10-25 | | Release date: | 2006-01-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of pyrimidine specificity in the MS2 RNA hairpin-coat-protein complex.
Rna, 7, 2001
|
|
2BU1
 
 | | MS2-RNA HAIRPIN (5BRU -5) COMPLEX | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *5BU*AP*CP*CP*CP*AP*UP*GP*U)-3', MS2 COAT PROTEIN | | Authors: | Grahn, E, Moss, T, Helgstrand, C, Fridborg, K, Sundaram, M, Tars, K, Lago, H, Stonehouse, N.J, Davis, D.R, Stockley, P.G, Liljas, L. | | Deposit date: | 2005-06-08 | | Release date: | 2005-08-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of pyrimidine specificity in the MS2 RNA hairpin-coat-protein complex.
Rna, 7, 2001
|
|
1BZ2
 
 | |
1BZT
 
 | |
1NJ3
 
 | | Structure and Ubiquitin Interactions of the Conserved NZF Domain of Npl4 | | Descriptor: | NPL4, ZINC ION | | Authors: | Wang, B, Alam, S.L, Meyer, H.H, Payne, M, Stemmler, T.L, Davis, D.R, Sundquist, W.I. | | Deposit date: | 2002-12-30 | | Release date: | 2003-04-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and ubiquitin interactions of the conserved zinc finger domain of Npl4.
J.Biol.Chem., 278, 2003
|
|
1M4P
 
 | | Structure of the Tsg101 UEV domain in complex with a HIV-1 PTAP "late domain" peptide, DYANA Ensemble | | Descriptor: | Gag Polyprotein, Tumor Susceptibility gene 101 protein | | Authors: | Pornillos, O, Alam, S.L, Davis, D.R, Sundquist, W.I. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein
Nat.Struct.Biol., 9, 2002
|
|
1M4Q
 
 | | STRUCTURE OF THE TSG101 UEV DOMAIN IN COMPLEX WITH A HIV-1 PTAP "LATE DOMAIN" PEPTIDE, CNS ENSEMBLE | | Descriptor: | Gag Polyprotein, Tumor Susceptibility gene 101 protein | | Authors: | Pornillos, O, Alam, S.L, Davis, D.R, Sundquist, W.I. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein
Nat.Struct.Biol., 9, 2002
|
|
1KPP
 
 | | Structure of the Tsg101 UEV domain | | Descriptor: | Tumor susceptibility gene 101 protein | | Authors: | Pornillos, O, Alam, S.L, Rich, R.L, Myszka, D.G, Davis, D.R, Sundquist, W.I. | | Deposit date: | 2002-01-02 | | Release date: | 2002-05-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and functional interactions of the Tsg101 UEV domain.
EMBO J., 21, 2002
|
|
1KPQ
 
 | | Structure of the Tsg101 UEV domain | | Descriptor: | Tumor susceptibility gene 101 protein | | Authors: | Pornillos, O, Alam, S.L, Rich, R.L, Myszka, D.G, Davis, D.R, Sundquist, W.I. | | Deposit date: | 2002-01-02 | | Release date: | 2002-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and functional interactions of the Tsg101 UEV domain.
EMBO J., 21, 2002
|
|
2KU0
 
 | | Inhibitor Induced Structural Change in the HCV IRES Domain IIa RNA | | Descriptor: | (7S)-7-[(dimethylamino)methyl]-1-[3-(dimethylamino)propyl]-7,8-dihydro-1H-furo[3,2-e]benzimidazol-2-amine, HCV IRES Domain IIa RNA | | Authors: | Paulsen, R.B, Seth, P.P, Swayze, E.E, Griffey, R.H, Skalicky, J.J, Cheatham III, T.E, Davis, D.R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Inhibitor-induced structural change in the HCV IRES domain IIa RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2LPT
 
 | |
2KTZ
 
 | | Inhibitor Induced Structural Change in the HCV IRES Domain IIa RNA | | Descriptor: | (7R)-7-[(dimethylamino)methyl]-1-[3-(dimethylamino)propyl]-7,8-dihydro-1H-furo[3,2-e]benzimidazol-2-amine, HCV IRES Domain IIa RNA | | Authors: | Paulsen, R.B, Seth, P.P, Swayze, E.E, Griffey, R.H, Skalicky, J.J, Cheatham III, T.E, Davis, D.R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Inhibitor-induced structural change in the HCV IRES domain IIa RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2LPS
 
 | |
1FL8
 
 | |
1BZU
 
 | |
1BZ3
 
 | |
4GNV
 
 | |
3GP3
 
 | |
3GW8
 
 | |
3GP5
 
 | |
