5Y6Q
 
 | |
1YSE
 
 | | Solution structure of the MAR-binding domain of SATB1 | | Descriptor: | DNA-binding protein SATB1 | | Authors: | Yamasaki, K, Yamaguchi, H. | | Deposit date: | 2005-02-08 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and DNA-binding Mode of the Matrix Attachment Region-binding Domain of the Transcription Factor SATB1 That Regulates the T-cell Maturation
J.Biol.Chem., 281, 2006
|
|
3A22
 
 | | Crystal Structure of beta-L-Arabinopyranosidase complexed with L-arabinose | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2009-04-27 | | Release date: | 2009-07-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A beta-l-Arabinopyranosidase from Streptomyces avermitilis is a novel member of glycoside hydrolase family 27.
J.Biol.Chem., 284, 2009
|
|
3A23
 
 | | Crystal Structure of beta-L-Arabinopyranosidase complexed with D-galactose | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2009-04-27 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A beta-l-Arabinopyranosidase from Streptomyces avermitilis is a novel member of glycoside hydrolase family 27.
J.Biol.Chem., 284, 2009
|
|
3A21
 
 | | Crystal Structure of Streptomyces avermitilis beta-L-Arabinopyranosidase | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2009-04-27 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A beta-l-Arabinopyranosidase from Streptomyces avermitilis is a novel member of glycoside hydrolase family 27.
J.Biol.Chem., 284, 2009
|
|
5JMT
 
 | | Crystal structure of Zika virus NS3 helicase | | Descriptor: | NS3 helicase | | Authors: | Tian, H, Ji, X, Yang, X, Xie, W, Yang, K, Chen, C, Wu, C, Chi, H, Mu, Z, Wang, Z, Yang, H. | | Deposit date: | 2016-04-29 | | Release date: | 2016-05-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | The crystal structure of Zika virus helicase: basis for antiviral drug design
Protein Cell, 7, 2016
|
|
1GSH
 
 | | STRUCTURE OF ESCHERICHIA COLI GLUTATHIONE SYNTHETASE AT PH 7.5 | | Descriptor: | GLUTATHIONE BIOSYNTHETIC LIGASE | | Authors: | Matsuda, K, Kato, H, Yamaguchi, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-05-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of glutathione synthetase at optimal pH: domain architecture and structural similarity with other proteins.
Protein Eng., 9, 1996
|
|
1HTJ
 
 | | STRUCTURE OF THE RGS-LIKE DOMAIN FROM PDZ-RHOGEF | | Descriptor: | KIAA0380 | | Authors: | Longenecker, K.L, Lewis, M.E, Chikumi, H, Gutkind, J.S, Derewenda, Z.S. | | Deposit date: | 2000-12-29 | | Release date: | 2001-07-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the RGS-like domain from PDZ-RhoGEF: linking heterotrimeric g protein-coupled signaling to Rho GTPases.
Structure, 9, 2001
|
|
1QZT
 
 | | Phosphotransacetylase from Methanosarcina thermophila | | Descriptor: | Phosphate acetyltransferase, SULFATE ION | | Authors: | Iyer, P.P, Lawrence, S.H, Luther, K.B, Rajashankar, K.R, Yennawar, H.P, Ferry, J.G, Schindelin, H. | | Deposit date: | 2003-09-17 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of phosphotransacetylase from the methanogenic archaeon Methanosarcina thermophila.
STRUCTURE, 12, 2004
|
|
5Y6M
 
 | | Zika virus helicase in complex with ADP-AlF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Helicase domain from Genome polyprotein, ... | | Authors: | Yang, X.Y, Chen, C, Tian, H.L, Chi, H, Mu, Z.Y, Zhang, T.Q, Yang, K.L, Zhao, Q, Liu, X.H, Wang, Z.F, Ji, X.Y, Yang, H.T. | | Deposit date: | 2017-08-12 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Mechanism of ATP hydrolysis by the Zika virus helicase.
FASEB J., 32, 2018
|
|
5Y6N
 
 | | Zika virus helicase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Helicase domain from Genome polyprotein, MANGANESE (II) ION | | Authors: | Yang, X.Y, Chen, C, Tian, H.L, Chi, H, Mu, Z.Y, Zhang, T.Q, Yang, K.L, Zhao, Q, Liu, X.H, Wang, Z.F, Ji, X.Y, Yang, H.T. | | Deposit date: | 2017-08-12 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Mechanism of ATP hydrolysis by the Zika virus helicase.
FASEB J., 32, 2018
|
|
2AF4
 
 | | Phosphotransacetylase from Methanosarcina thermophila co-crystallized with coenzyme A | | Descriptor: | COENZYME A, Phosphate acetyltransferase | | Authors: | Lawrence, S.H, Luther, K.B, Ferry, J.G, Schindelin, H. | | Deposit date: | 2005-07-25 | | Release date: | 2006-01-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Structural and functional studies suggest a catalytic mechanism for the phosphotransacetylase from Methanosarcina thermophila.
J.Bacteriol., 188, 2006
|
|
2AF3
 
 | | Phosphotransacetylase from Methanosarcina thermophila soaked with Coenzyme A | | Descriptor: | COENZYME A, Phosphate acetyltransferase, SULFATE ION | | Authors: | Lawrence, S.H, Luther, K.B, Ferry, J.G, Schindelin, H. | | Deposit date: | 2005-07-25 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional studies suggest a catalytic mechanism for the phosphotransacetylase from Methanosarcina thermophila.
J.Bacteriol., 188, 2006
|
|
4V98
 
 | | The 8S snRNP Assembly Intermediate | | Descriptor: | CG10419, Icln, LD23602p, ... | | Authors: | Grimm, C, Pelz, J.P, Schindelin, H, Diederichs, K, Kuper, J, Kisker, C. | | Deposit date: | 2012-05-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of Assembly Chaperone- Mediated snRNP Formation.
Mol.Cell, 49, 2013
|
|
8H4E
 
 | | Blasnase-T13A/P55N with D-asn | | Descriptor: | D-ASPARAGINE, FORMIC ACID, L-asparaginase, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H46
 
 | | Blasnase-T13A/P55N with L-asn | | Descriptor: | ASPARAGINE, FORMIC ACID, GLYCEROL, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H47
 
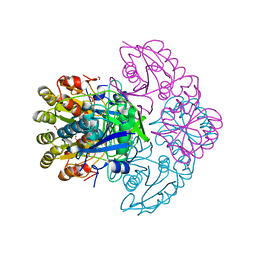 | | Blasnase-T13A/P55F | | Descriptor: | FORMIC ACID, GLYCEROL, L-asparaginase, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H4B
 
 | | Blasnase-T13A/M57P with L-asn | | Descriptor: | ASPARAGINE, FORMIC ACID, L-asparaginase, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H4D
 
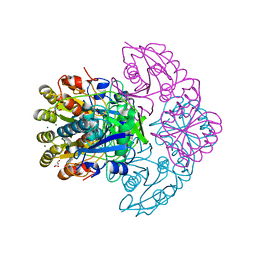 | | Blasnase-T13A/M57N | | Descriptor: | FORMIC ACID, GLYCEROL, L-asparaginase, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H44
 
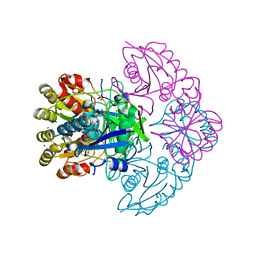 | | Blasnase-P55N | | Descriptor: | FORMIC ACID, L-asparaginase, MAGNESIUM ION | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H45
 
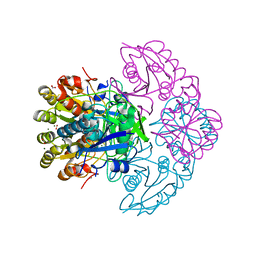 | | Blasnase-T13A/P55N | | Descriptor: | FORMIC ACID, L-asparaginase, MAGNESIUM ION | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H48
 
 | | Blasnase-T13A/P55F with L-asn | | Descriptor: | ASPARAGINE, FORMIC ACID, L-asparaginase, ... | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H49
 
 | | Blasnase-M57P | | Descriptor: | FORMIC ACID, L-asparaginase, MAGNESIUM ION | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H4A
 
 | | Blasnase-T13A/M57P | | Descriptor: | FORMIC ACID, L-asparaginase, MAGNESIUM ION | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
8H4C
 
 | | Blasnase-T13A/M57P | | Descriptor: | FORMIC ACID, L-asparaginase, MAGNESIUM ION | | Authors: | Lu, F, Wang, W, Chi, H, Ran, T. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based rational design of Bacillus licheniformis L-asparaginase with low/no D-asparaginase activity for a safer enzyme
To Be Published
|
|
