8XSF
 
 | | SARS-CoV-2 RBD + IMCAS-364 + hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, IMCAS-364 H chain, ... | | Authors: | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | Deposit date: | 2024-01-09 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8Y0Y
 
 | | Cryo-EM structure of the 123-316 scDb/PT-RBD complex | | Descriptor: | 123-316 scDb, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Jia, G.W, Tong, Z, Tong, J.Y, Su, Z.M. | | Deposit date: | 2024-01-23 | | Release date: | 2024-07-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XSJ
 
 | | SARS-CoV-2 Omicron BA.4 RBD + IMCAS-316 + ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | Deposit date: | 2024-01-09 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XSL
 
 | | SARS-CoV-2 spike + IMCAS-123 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS-123 heavy chain, ... | | Authors: | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | Deposit date: | 2024-01-09 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XSI
 
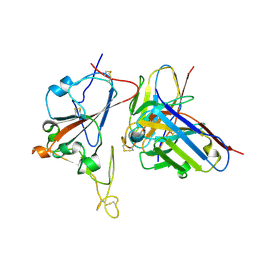 | | SARS-CoV-2 RBD + IMCAS-364 (Local Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS-364 H chain, IMCAS-364 L chain, ... | | Authors: | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | Deposit date: | 2024-01-09 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
8XSE
 
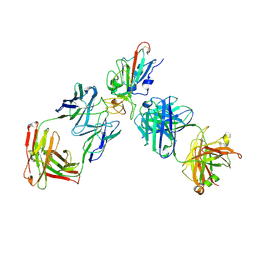 | | SARS-CoV-2 RBD + IMCAS-123 + IMCAS-72 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS-123 H chain, IMCAS-123 L chain, ... | | Authors: | Tong, Z, Cui, Y, Xie, Y, Tong, J, Gao, G.F, Qi, J. | | Deposit date: | 2024-01-09 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Deciphering a reliable synergistic bispecific strategy of rescuing antibodies for SARS-CoV-2 escape variants, including BA.2.86, EG.5.1, and JN.1.
Cell Rep, 43, 2024
|
|
6N05
 
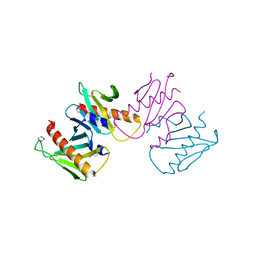 | | Structure of anti-crispr protein, AcrIIC2 | | Descriptor: | AcrIIC2 | | Authors: | Shah, M, Thavalingham, A, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2018-11-06 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2.
Nat Commun, 10, 2019
|
|
8HOG
 
 | | Crystal structure of Bcl-2 in complex with sonrotoclax | | Descriptor: | Apoptosis regulator Bcl-2, ~{N}-[4-[(4-methyl-4-oxidanyl-cyclohexyl)methylamino]-3-nitro-phenyl]sulfonyl-4-[2-[(2~{S})-2-(2-propan-2-ylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide | | Authors: | Liu, J, Xu, M, Feng, Y, Hong, Y, Liu, Y. | | Deposit date: | 2022-12-10 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy.
Blood, 143, 2024
|
|
8HOH
 
 | | Crystal structure of Bcl-2 G101V in complex with sonrotoclax | | Descriptor: | Apoptosis regulator Bcl-2, ~{N}-[4-[(4-methyl-4-oxidanyl-cyclohexyl)methylamino]-3-nitro-phenyl]sulfonyl-4-[2-[(2~{S})-2-(2-propan-2-ylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide | | Authors: | Liu, J, Xu, M, Feng, Y, Hong, Y, Liu, Y. | | Deposit date: | 2022-12-10 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy.
Blood, 143, 2024
|
|
8HOI
 
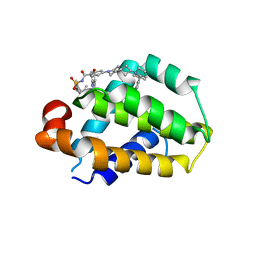 | | Crystal structure of Bcl-2 D103Y in complex with sonrotoclax | | Descriptor: | Apoptosis regulator Bcl-2, FORMIC ACID, ~{N}-[4-[(4-methyl-4-oxidanyl-cyclohexyl)methylamino]-3-nitro-phenyl]sulfonyl-4-[2-[(2~{S})-2-(2-propan-2-ylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide | | Authors: | Liu, J, Xu, M, Feng, Y, Hong, Y, Liu, Y. | | Deposit date: | 2022-12-10 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Sonrotoclax overcomes BCL2 G101V mutation-induced venetoclax resistance in preclinical models of hematologic malignancy.
Blood, 143, 2024
|
|
8YLC
 
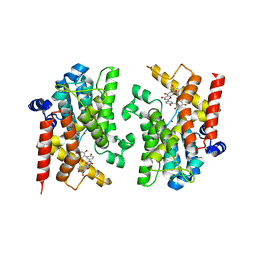 | | The crystal structure of PDE4D with Amentoflavone | | Descriptor: | 3',5'-cyclic-AMP phosphodiesterase 4D, 8-[5-[5,7-bis(oxidanyl)-4-oxidanylidene-chromen-2-yl]-2-oxidanyl-phenyl]-2-(4-hydroxyphenyl)-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | Authors: | Huang, Y.-Y, Luo, H.-B. | | Deposit date: | 2024-03-06 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.30003715 Å) | | Cite: | Discovery of amentoflavone as a natural PDE4 inhibitor with anti-fibrotic effects
Chin.Chem.Lett., 2024
|
|
7CRQ
 
 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (2:1 binding mode) | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRP
 
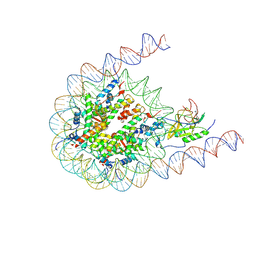 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (1:1 binding mode) | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRO
 
 | | NSD2 bearing E1099K/T1150A dual mutation in complex with 187-bp NCP | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRR
 
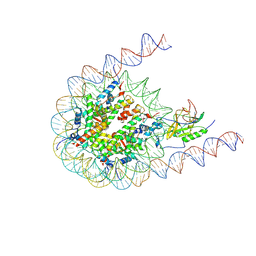 | | Native NSD3 bound to 187-bp nucleosome | | Descriptor: | DNA (168-MER), DNA(168-MER), Histone H2A, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
8HJB
 
 | | Crystal structure of Pseudomonas aeruginosa PvrA with coenzyme A | | Descriptor: | COENZYME A, TetR family transcriptional regulator | | Authors: | Liang, H, Bartlam, M. | | Deposit date: | 2022-11-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
7Y0Y
 
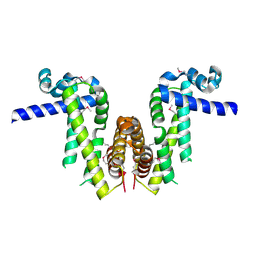 | | Crystal structure of Pseudomonas aeruginosa PvrA (SeMet) | | Descriptor: | TetR family transcriptional regulator | | Authors: | Liang, H, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
7YC9
 
 | | Co-crystal structure of BTK kinase domain with inhibitor | | Descriptor: | (7~{S})-2-(4-bromanyl-3,5-dimethoxy-phenyl)-7-(1-propanoylpiperidin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, 1,2-ETHANEDIOL, Tyrosine-protein kinase BTK | | Authors: | Zhou, X. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of BGB-8035, a Highly Selective Covalent Inhibitor of Bruton's Tyrosine Kinase for B-Cell Malignancies and Autoimmune Diseases.
J.Med.Chem., 66, 2023
|
|
7Y0Z
 
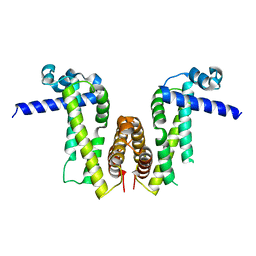 | | Crystal structure of Pseudomonas aeruginosa PvrA | | Descriptor: | TetR family transcriptional regulator | | Authors: | Liang, H, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
7BV7
 
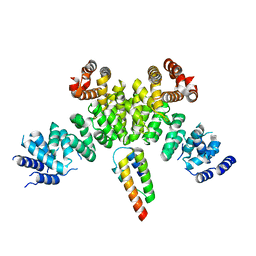 | | INTS3 complexed with INTS6 | | Descriptor: | Integrator complex subunit 3, Integrator complex subunit 6 | | Authors: | Jia, Y, Bharath, S.R, Song, H. | | Deposit date: | 2020-04-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the INTS3/INTS6 complex reveals the functional importance of INTS3 dimerization in DSB repair.
Cell Discov, 7, 2021
|
|
8HJ4
 
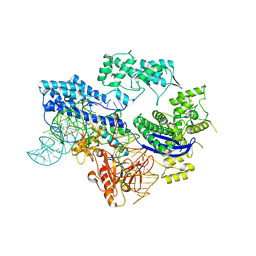 | |
8JFU
 
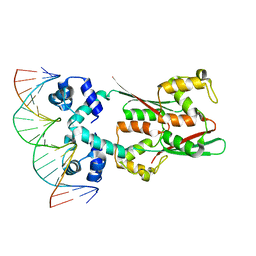 | | AcrIIA15 in complex with palindromic DNA substrate | | Descriptor: | AcrIIA15, DNA (5'-D(*AP*TP*TP*AP*TP*GP*AP*CP*AP*AP*AP*TP*GP*TP*CP*AP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*TP*GP*AP*CP*AP*TP*TP*TP*GP*TP*CP*AP*TP*AP*A)-3') | | Authors: | Deng, X, Wang, Y. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | An anti-CRISPR that represses its own transcription while blocking Cas9-target DNA binding.
Nat Commun, 15, 2024
|
|
8JFO
 
 | |
8JFT
 
 | | Cryo-EM structure of SaCas9-AcrIIA15 CTD-sgRNA complex | | Descriptor: | AcrIIA15, CRISPR-associated endonuclease Cas9, sgRNA of SaCas9 | | Authors: | Deng, X, Wang, Y. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | An anti-CRISPR that represses its own transcription while blocking Cas9-target DNA binding.
Nat Commun, 15, 2024
|
|
8JFR
 
 | | N-terminal domain of AcrIIA15 in complex with palindromic DNA substrate | | Descriptor: | AcrIIA15, DNA (5'-D(*AP*TP*TP*AP*TP*GP*AP*CP*AP*AP*AP*TP*GP*TP*CP*AP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*TP*GP*AP*CP*AP*TP*TP*TP*GP*TP*CP*AP*TP*AP*A)-3') | | Authors: | Deng, X, Wang, Y. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An anti-CRISPR that represses its own transcription while blocking Cas9-target DNA binding.
Nat Commun, 15, 2024
|
|
