7ASQ
 
 | |
7OQM
 
 | | Human OMPD-domain of UMPS in complex with substrate OMP at 1.05 Angstroms resolution, 20 minutes soaking | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OQI
 
 | | Human OMPD-domain of UMPS in complex with substrate OMP at 1.15 Angstrom resolution, 10 minutes soaking | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OQK
 
 | | Human OMPD-domain of UMPS in complex with substrate OMP at 1.10 Angstroms resolution, 15 minutes soaking | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OQN
 
 | | Human OMPD-domain of UMPS in complex with substrate OMP at 1.10 Angstroms resolution, 30 minutes soaking | | Descriptor: | GLYCEROL, SULFATE ION, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OQF
 
 | | Human OMPD-domain of UMPS in complex with OMP at 1.05 Angstrom resolution, 5 minutes soaking | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OUZ
 
 | |
7OV0
 
 | |
7OTU
 
 | | Human OMPD-domain of UMPS in complex with 6-hydroxy-UMP at 0.95 Angstroms resolution, crystal 2 | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Isoform 2 of Uridine 5'-monophosphate synthase | | Authors: | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | Deposit date: | 2021-06-10 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7Q1H
 
 | |
4NOX
 
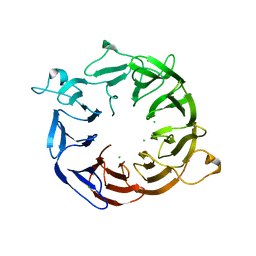 | | Structure of the nine-bladed beta-propeller of eIF3b | | Descriptor: | CHLORIDE ION, Eukaryotic translation initiation factor 3 subunit B | | Authors: | Liu, Y, Neumann, P, Kuhle, B, Monecke, T, Ficner, R. | | Deposit date: | 2013-11-20 | | Release date: | 2014-09-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.722 Å) | | Cite: | Translation initiation factor eIF3b contains a nine-bladed beta-propeller and interacts with the 40S ribosomal subunit
Structure, 22, 2014
|
|
6H60
 
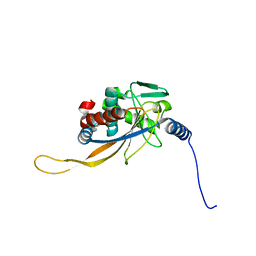 | | pseudo-atomic structural model of the E3BP component of the human pyruvate dehydrogenase multienzyme complex | | Descriptor: | Pyruvate dehydrogenase protein X component, mitochondrial | | Authors: | Haselbach, D, Prajapati, S, Tittmann, K, Stark, H. | | Deposit date: | 2018-07-25 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural and Functional Analyses of the Human PDH Complex Suggest a "Division-of-Labor" Mechanism by Local E1 and E3 Clusters.
Structure, 27, 2019
|
|
6H55
 
 | | core of the human pyruvate dehydrogenase (E2) | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | | Authors: | Haselbach, D, Prajapati, S, Tittmann, K, Stark, H. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural and Functional Analyses of the Human PDH Complex Suggest a "Division-of-Labor" Mechanism by Local E1 and E3 Clusters.
Structure, 27, 2019
|
|
1IGD
 
 | |
1IGC
 
 | |
1PGB
 
 | |
1PGA
 
 | |
8OMV
 
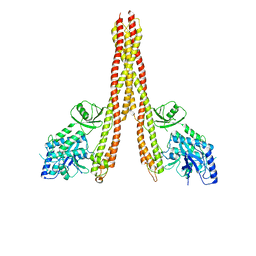 | | Crystal structure of the constitutively active S117E/S181E mutant of human IKK2 | | Descriptor: | Inhibitor of nuclear factor kappa-B kinase subunit beta | | Authors: | McEwen, A.G, Li, C, Moro, S, Poussin-Courmontagne, P, Zanier, K. | | Deposit date: | 2023-03-31 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.16 Å) | | Cite: | Molecular mechanism of IKK catalytic dimer docking to NF-kappa B substrates.
Nat Commun, 15, 2024
|
|
1LY3
 
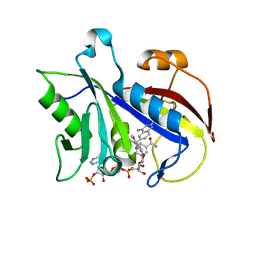 | | ANALYSIS OF QUINAZOLINE AND PYRIDOPYRIMIDINE N9-C10 REVERSED BRIDGE ANTIFOLATES IN COMPLEX WITH NADP+ AND PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE | | Descriptor: | 2,4-DIAMINO-6-[N-(2',5'-DIMETHOXYBENZYL)-N-METHYLAMINO]QUINAZOLINE, DIHYDROFOLATE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Queener, S.F, Gangjee, A. | | Deposit date: | 2002-06-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of quinazoline and pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1DAJ
 
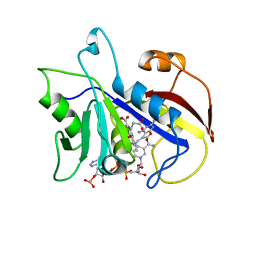 | | COMPARISON OF TERNARY COMPLEXES OF PNEUMOCYSTIS CARINII AND WILD TYPE HUMAN DIHYDROFOLATE REDUCTASE WITH COENZYME NADPH AND A NOVEL CLASSICAL ANTITUMOR FURO[2,3D]PYRIMIDINE ANTIFOLATE | | Descriptor: | DIHYDROFOLATE REDUCTASE, N-[4-[(2,4-DIAMINOFURO[2,3D]PYRIMIDIN-5-YL)METHYL]METHYLAMINO]-BENZOYL]-L-GLUTAMATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Gangjee, A, Devraj, R, Queener, S.F, Blakley, R.L. | | Deposit date: | 1997-07-29 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Comparison of ternary complexes of Pneumocystis carinii and wild-type human dihydrofolate reductase with coenzyme NADPH and a novel classical antitumor furo[2,3-d]pyrimidine antifolate.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1LY4
 
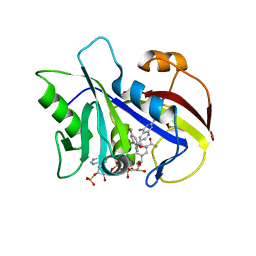 | | Analysis of quinazoline and PYRIDO[2,3D]PYRIMIDINE N9-C10 reversed bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase | | Descriptor: | 2,4-DIAMINO-6-[N-(3',5'-DIMETHOXYBENZYL)-N-METHYLAMINO]PYRIDO[2,3-D]PYRIMIDINE, DIHYDROFOLATE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Queener, S.F, Gangjee, A. | | Deposit date: | 2002-06-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of quinazoline and pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1MPE
 
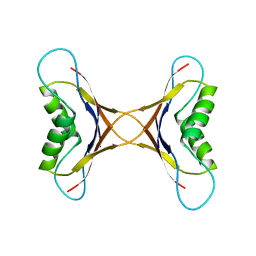 | |
1MVK
 
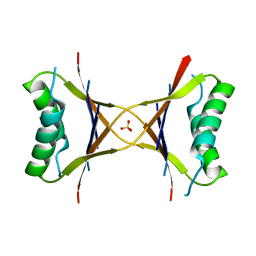 | | X-ray structure of the tetrameric mutant of the B1 domain of streptococcal protein G | | Descriptor: | Immunoglobulin G binding protein G, SULFATE ION | | Authors: | Frank, M.K, Dyda, F, Dobrodumov, A, Gronenborn, A.M. | | Deposit date: | 2002-09-25 | | Release date: | 2002-10-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Core mutations switch monomeric protein GB1 into an intertwined tetramer.
Nat.Struct.Biol., 9, 2002
|
|
3GB1
 
 | | STRUCTURES OF B1 DOMAIN OF STREPTOCOCCAL PROTEIN G | | Descriptor: | PROTEIN (B1 DOMAIN OF STREPTOCOCCAL PROTEIN G) | | Authors: | Clore, G.M. | | Deposit date: | 1999-05-02 | | Release date: | 1999-06-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Improving the Packing and Accuracy of NMR Structures with a Pseudopotential for the Radius of Gyration
J.Am.Chem.Soc., 121, 1999
|
|
1Q10
 
 | |
