5Y5J
 
 | | Time-resolved SFX structure of cytochrome P450nor: dark-2 data in the presence of NADH (resting state) | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5K
 
 | | Time-resolved SFX structure of cytochrome P450nor : 20 ms after photo-irradiation of caged NO in the absence of NADH (NO-bound state), light data | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
3NKM
 
 | | Crystal structure of mouse autotaxin | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NKN
 
 | | Crystal structure of mouse autotaxin in complex with 14:0-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NKQ
 
 | | Crystal structure of mouse autotaxin in complex with 18:3-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E,12Z,15Z)-octadeca-9,12,15-trienoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NKO
 
 | | Crystal structure of mouse autotaxin in complex with 16:0-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NKP
 
 | | Crystal structure of mouse autotaxin in complex with 18:1-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NKR
 
 | | Crystal structure of mouse autotaxin in complex with 22:6-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (4Z,7E,10E,13Z,16Z,19Z)-docosa-4,7,10,13,16,19-hexaenoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
3WAV
 
 | | Crystal Structure of Autotaxin in Complex with Compound 10 | | Descriptor: | (5Z)-5-(3,4-dichlorobenzylidene)-2-(4-methylpiperazin-1-yl)-1,3-thiazol-4(5H)-one, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-09 | | Release date: | 2013-07-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Screening and X-ray Crystal Structure-based Optimization of Autotaxin (ENPP2) Inhibitors, Using a Newly Developed Fluorescence Probe
Acs Chem.Biol., 8, 2013
|
|
2DB3
 
 | | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ATP-dependent RNA helicase vasa, MAGNESIUM ION, ... | | Authors: | Sengoku, T, Nureki, O, Nakamura, A, Kobayashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-14 | | Release date: | 2006-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa.
Cell(Cambridge,Mass.), 125, 2006
|
|
3WAY
 
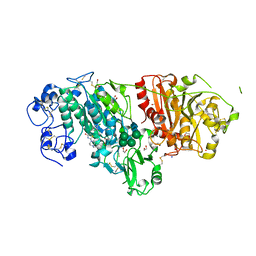 | | Crystal Structure of Autotaxin in Complex with 4BoA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-09 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Screening and X-ray Crystal Structure-based Optimization of Autotaxin (ENPP2) Inhibitors, Using a Newly Developed Fluorescence Probe
Acs Chem.Biol., 8, 2013
|
|
3X3B
 
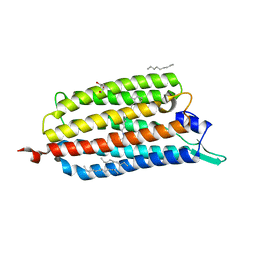 | | Crystal structure of the light-driven sodium pump KR2 in acidic state | | Descriptor: | DI(HYDROXYETHYL)ETHER, OLEIC ACID, RETINAL, ... | | Authors: | Kato, H.E, Inoue, K, Abe-Yoshizumi, R, Kato, Y, Ono, H, Konno, M, Ishizuka, T, Hoque, M.R, Hososhima, S, Kunitomo, H, Ito, J, Yoshizawa, S, Yamashita, K, Takemoto, M, Nishizawa, T, Taniguchi, R, Kogure, K, Maturana, A.D, Iino, Y, Yawo, H, Ishitani, R, Kandori, H, Nureki, O. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for Na(+) transport mechanism by a light-driven Na(+) pump
Nature, 521, 2015
|
|
3X3C
 
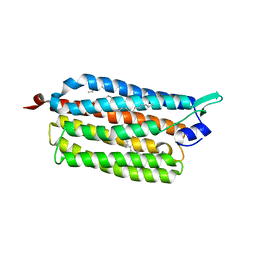 | | Crystal structure of the light-driven sodium pump KR2 in neutral state | | Descriptor: | OLEIC ACID, RETINAL, Sodium pumping rhodopsin | | Authors: | Kato, H.E, Inoue, K, Abe-Yoshizumi, R, Kato, Y, Ono, H, Konno, M, Ishizuka, T, Hoque, M.R, Hososhima, S, Kunitomo, H, Ito, J, Yoshizawa, S, Yamashita, K, Takemoto, M, Nishizawa, T, Taniguchi, R, Kogure, K, Maturana, A.D, Iino, Y, Yawo, H, Ishitani, R, Kandori, H, Nureki, O. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for Na(+) transport mechanism by a light-driven Na(+) pump
Nature, 521, 2015
|
|
6LMX
 
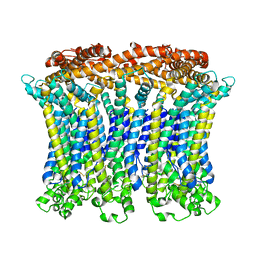 | | Cryo-EM structure of the CALHM chimeric construct (9-mer) | | Descriptor: | Calcium homeostasis modulator 1,Calcium homeostasis modulator protein 2 | | Authors: | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-29 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
6LMV
 
 | | Cryo-EM structure of the C. elegans CLHM-1 | | Descriptor: | Calcium homeostasis modulator protein | | Authors: | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
6LRY
 
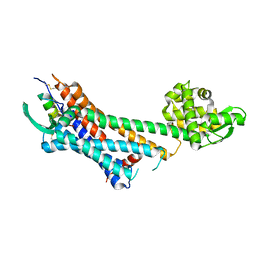 | | Crystal structure of human endothelin ETB receptor in complex with sarafotoxin S6b | | Descriptor: | Endothelin receptor type B,Endolysin,Endothelin receptor type B, Sarafotoxin-B | | Authors: | Izume, T, Miyauchi, H, Shihoya, W, Nureki, O. | | Deposit date: | 2020-01-16 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human endothelin ETBreceptor in complex with sarafotoxin S6b.
Biochem.Biophys.Res.Commun., 528, 2020
|
|
6LMW
 
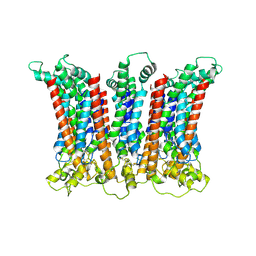 | | Cryo-EM structure of the CALHM chimeric construct (8-mer) | | Descriptor: | Calcium homeostasis modulator 1,Calcium homeostasis modulator protein 2 | | Authors: | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
6LPJ
 
 | | A2AR crystallized in EROCOC17+4, LCP-SFX at 277 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
6LPL
 
 | | A2AR crystallized in EROCOC17+4, SS-ROX at 100 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-11 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
8H1J
 
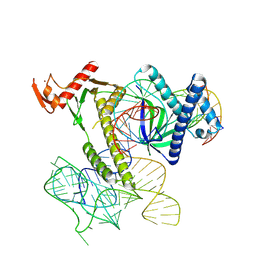 | | Cryo-EM structure of the TnpB-omegaRNA-target DNA ternary complex | | Descriptor: | Non-target strand, RNA-guided DNA endonuclease TnpB, Target strand, ... | | Authors: | Nakagawa, R, Hirano, H, Omura, S, Nureki, O. | | Deposit date: | 2022-10-03 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of the transposon-associated TnpB enzyme.
Nature, 616, 2023
|
|
4GEL
 
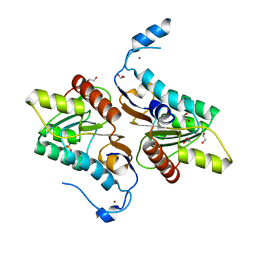 | | Crystal structure of Zucchini | | Descriptor: | 1,2-ETHANEDIOL, Mitochondrial cardiolipin hydrolase, PHOSPHATE ION, ... | | Authors: | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
8GW8
 
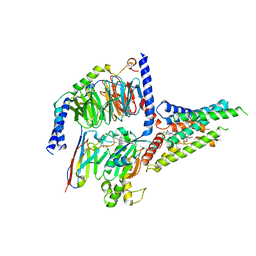 | | the human PTH1 receptor bound to an intracellular biased agonist | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform Gnas-2 of Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Okamoto, H.H, Nureki, O. | | Deposit date: | 2022-09-16 | | Release date: | 2023-06-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Class B1 GPCR activation by an intracellular agonist.
Nature, 618, 2023
|
|
4GEN
 
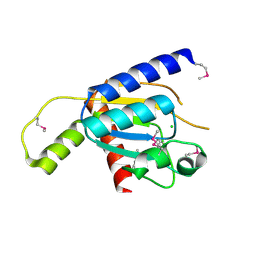 | | Crystal structure of Zucchini (monomer) | | Descriptor: | CHLORIDE ION, Mitochondrial cardiolipin hydrolase | | Authors: | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
8HIO
 
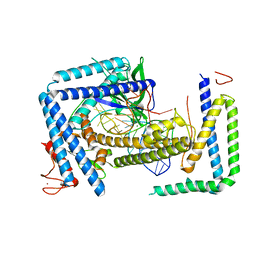 | | Cryo-EM structure of the Cas12m2-crRNA binary complex | | Descriptor: | Cas12m2, MAGNESIUM ION, RNA (56-MER), ... | | Authors: | Omura, N.S, Nakagawa, R, Wu, Y.W, Sudfeld, C, Warren, V.R, Hirano, H, Kusakizako, T, Kise, Y, Lebbink, H.G.J, Itoh, Y, Oost, V.D.J, Nureki, O. | | Deposit date: | 2022-11-21 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Mechanistic and evolutionary insights into a type V-M CRISPR-Cas effector enzyme.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8HHM
 
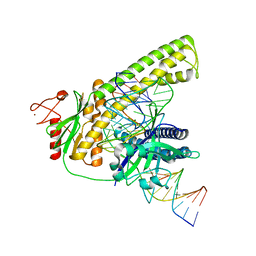 | | Cryo-EM structure of the Cas12m2-crRNA-target DNA ternary complex intermediate state | | Descriptor: | Cas12m2, DNA (36-MER), MAGNESIUM ION, ... | | Authors: | Omura, N.S, Nakagawa, R, Wu, Y.W, Sudfeld, C, Warren, V.R, Hirano, H, Kusakizako, T, Kise, Y, Lebbink, H.G.J, Itoh, Y, Oost, V.D.J, Nureki, O. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Mechanistic and evolutionary insights into a type V-M CRISPR-Cas effector enzyme.
Nat.Struct.Mol.Biol., 30, 2023
|
|
