3H0N
 
 | |
3FKQ
 
 | |
3HBZ
 
 | |
3GO5
 
 | |
3H41
 
 | |
3H50
 
 | |
3IRB
 
 | |
6CNV
 
 | | INFLUENZA B/BRISBANE HEMAGGLUTININ FAB CR9115 SD84H COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR9114 Fab heavy chain, ... | | Authors: | Luo, J, Obmolova, G. | | Deposit date: | 2018-03-09 | | Release date: | 2018-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin.
Science, 362, 2018
|
|
8SXP
 
 | |
6CNW
 
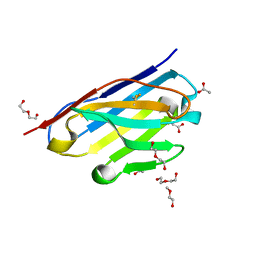 | | STRUCTURE OF HUMANIZED SINGLE DOMAIN ANTIBODY SD84 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, humanized antibody SD84h | | Authors: | Luo, J, Obmolova, O. | | Deposit date: | 2018-03-09 | | Release date: | 2018-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin.
Science, 362, 2018
|
|
8D0Z
 
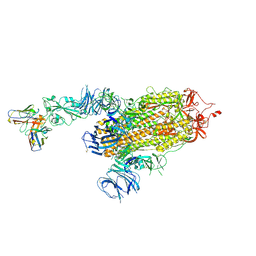 | | S728-1157 IgG in complex with SARS-CoV-2-6P-Mut7 Spike protein (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S728-1157 Fab heavy chain variable region, S728-1157 Fab light chain variable region, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2022-05-26 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Site of vulnerability on SARS-CoV-2 spike induces broadly protective antibody against antigenically distinct Omicron subvariants.
J.Clin.Invest., 133, 2023
|
|
2RNK
 
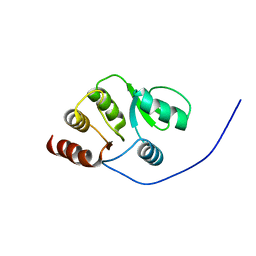 | | NMR structure of the domain 513-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B.W, Wilson, I.A, Stevens, R.C, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-11 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
2VIU
 
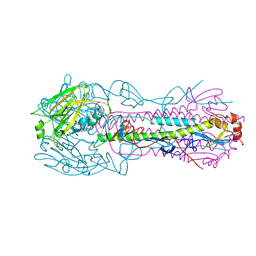 | | INFLUENZA VIRUS HEMAGGLUTININ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bizebard, T, Fleury, D, Gigant, B, Wharton, S.A, Skehel, J.J, Knossow, M. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antigen distortion allows influenza virus to escape neutralization.
Nat.Struct.Biol., 5, 1998
|
|
2VIS
 
 | | INFLUENZA VIRUS HEMAGGLUTININ, (ESCAPE) MUTANT WITH THR 131 REPLACED BY ILE, COMPLEXED WITH A NEUTRALIZING ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, IMMUNOGLOBULIN (IGG1, ... | | Authors: | Bizebard, T, Fleury, D, Gigant, B, Wharton, S.A, Skehel, J.J, Knossow, M. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Antigen distortion allows influenza virus to escape neutralization.
Nat.Struct.Biol., 5, 1998
|
|
2VIT
 
 | | INFLUENZA VIRUS HEMAGGLUTININ, MUTANT WITH THR 155 REPLACED BY ILE, COMPLEXED WITH A NEUTRALIZING ANTIBODY | | Descriptor: | HEMAGGLUTININ, IMMUNOGLOBULIN (IGG1, LAMBDA), ... | | Authors: | Bizebard, T, Fleury, D, Gigant, B, Wharton, S.A, Skehel, J.J, Knossow, M. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-29 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Antigen distortion allows influenza virus to escape neutralization.
Nat.Struct.Biol., 5, 1998
|
|
2VIR
 
 | | INFLUENZA VIRUS HEMAGGLUTININ COMPLEXED WITH A NEUTRALIZING ANTIBODY | | Descriptor: | HEMAGGLUTININ, IMMUNOGLOBULIN (IGG1, LAMBDA), ... | | Authors: | Bizebard, T, Fleury, D, Gigant, B, Wharton, S.A, Skehel, J.J, Knossow, M. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Antigen distortion allows influenza virus to escape neutralization.
Nat.Struct.Biol., 5, 1998
|
|
5J75
 
 | | Fluorogen Activating Protein AM2.2 in complex with ML342 | | Descriptor: | N,4-dimethyl-N-{2-oxo-2-[4-(pyridin-2-yl)piperazin-1-yl]ethyl}benzene-1-sulfonamide, PHOSPHATE ION, scFv AM2.2 | | Authors: | Stanfield, R.L, Wilson, I.A, Wu, Y. | | Deposit date: | 2016-04-05 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Small-Molecule Nonfluorescent Inhibitors of Fluorogen-Fluorogen Activating Protein Binding Pair.
J Biomol Screen, 21, 2016
|
|
3OZ2
 
 | |
3PAY
 
 | |
3PL0
 
 | |
3RJ0
 
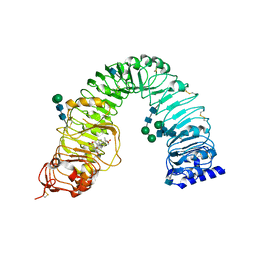 | | Plant steroid receptor BRI1 ectodomain in complex with brassinolide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Brassinolide, Protein BRASSINOSTEROID INSENSITIVE 1, ... | | Authors: | Hothorn, M. | | Deposit date: | 2011-04-14 | | Release date: | 2011-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | Structural basis of steroid hormone perception by the receptor kinase BRI1.
Nature, 474, 2011
|
|
3R4R
 
 | |
3O0F
 
 | |
2YPI
 
 | |
3OHG
 
 | |
