1KV6
 
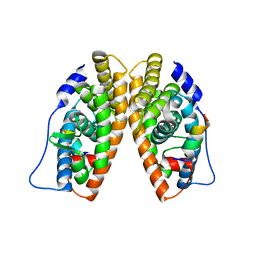 | | X-ray structure of the orphan nuclear receptor ERR3 ligand-binding domain in the constitutively active conformation | | Descriptor: | ESTROGEN-RELATED RECEPTOR GAMMA, steroid receptor coactivator 1 | | Authors: | Greschik, H, Wurtz, J.-M, Sanglier, S, Bourguet, W, van Dorsselaer, A, Moras, D, Renaud, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-01-25 | | Release date: | 2003-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Evidence for Ligand-Independent Transcriptional Activation by the Estrogen-Related Receptor 3
Mol.Cell, 9, 2002
|
|
7K7Q
 
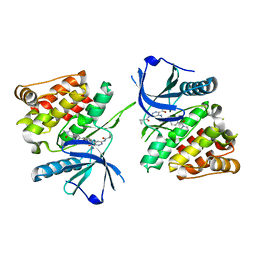 | |
7K7O
 
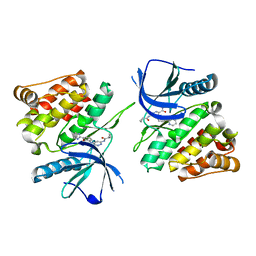 | |
3QWR
 
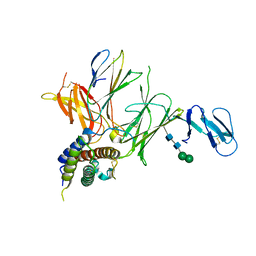 | | Crystal structure of IL-23 in complex with an adnectin | | Descriptor: | ADNECTIN, Interleukin-12 subunit beta, Interleukin-23 subunit alpha, ... | | Authors: | Wei, A, Sheriff, S. | | Deposit date: | 2011-02-28 | | Release date: | 2012-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structures of adnectin/protein complexes reveal an expanded binding footprint.
Structure, 20, 2012
|
|
8K36
 
 | |
8K35
 
 | | Structure of the bacteriophage lambda tail tip complex | | Descriptor: | IRON/SULFUR CLUSTER, Tail tip assembly protein I, Tail tip protein L, ... | | Authors: | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
8K37
 
 | | Structure of the bacteriophage lambda neck | | Descriptor: | Head-tail connector protein FII, Tail tube protein, Tail tube terminator protein | | Authors: | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
8K38
 
 | | The structure of bacteriophage lambda portal-adaptor | | Descriptor: | Head completion protein, Portal protein B | | Authors: | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
8K39
 
 | | Structure of the bacteriophage lambda portal vertex | | Descriptor: | Major capsid protein, Portal protein B | | Authors: | Xiao, H, Tan, L, Cheng, L.P, Liu, H.R. | | Deposit date: | 2023-07-14 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the siphophage neck-Tail complex suggests that conserved tail tip proteins facilitate receptor binding and tail assembly.
Plos Biol., 21, 2023
|
|
3QWQ
 
 | |
6NSL
 
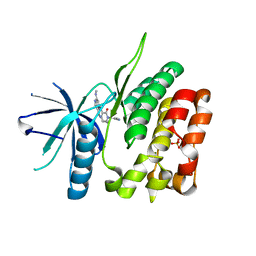 | | CRYSTAL STRUCTURE OF TYROSINE KINASE 2 JH2 (PSEUDO KINASE DOMAIN) COMPLEXED WITH Compound-6c AKA 6-((1-(4-CYANOPHENY L)-2-OXO-1,2-DIHYDRO-3-PYRIDINYL)AMINO)-N-CYCLOPROPYL-8-(M ETHYLAMINO)IMIDAZO[1,2-B]PYRIDAZINE-3-CARBOXAMIDE | | Descriptor: | 6-{[1-(4-cyanophenyl)-2-oxo-1,2-dihydropyridin-3-yl]amino}-N-cyclopropyl-8-(methylamino)imidazo[1,2-b]pyridazine-3-carboxamide, Non-receptor tyrosine-protein kinase TYK2, SULFATE ION | | Authors: | Muckelbauer, J.M, Khan, J.A. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of Imidazo[1,2-b]pyridazine Derivatives as Potent, Selective, and Orally Active Tyk2 JH2 Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6NZR
 
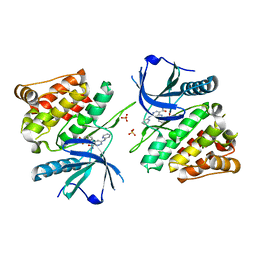 | |
6NZQ
 
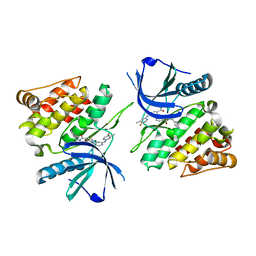 | |
6NZF
 
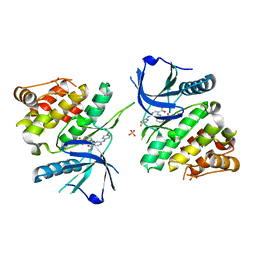 | |
6NZH
 
 | |
6NZE
 
 | |
6NZP
 
 | |
6JOY
 
 | | The X-ray Crystallographic Structure of Branching Enzyme from Rhodothermus obamensis STB05 | | Descriptor: | 1,4-alpha-glucan branching enzyme GlgB | | Authors: | Li, Z.F, Ban, X.F, Jiang, H.M, Wang, Z, Jin, T.C, Li, C.M, Gu, Z.B. | | Deposit date: | 2019-03-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Flexible Loop in Carbohydrate-Binding Module 48 Allosterically Modulates Substrate Binding of the 1,4-alpha-Glucan Branching Enzyme.
J.Agric.Food Chem., 69, 2021
|
|
4NWM
 
 | |
3H3B
 
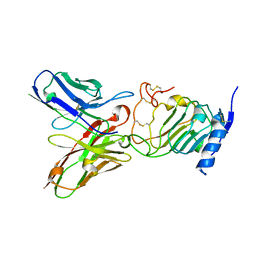 | | Crystal structure of the single-chain Fv (scFv) fragment of an anti-ErbB2 antibody chA21 in complex with residues 1-192 of ErbB2 extracellular domain | | Descriptor: | Receptor tyrosine-protein kinase erbB-2, anti-ErbB2 antibody chA21 | | Authors: | Zhou, H, Liu, Y, Niu, L, Zhu, J, Teng, M. | | Deposit date: | 2009-04-16 | | Release date: | 2010-04-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Insights into the Down-regulation of Overexpressed p185her2/neu Protein of Transformed Cells by the Antibody chA21.
J.Biol.Chem., 286, 2011
|
|
6L35
 
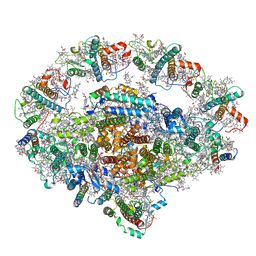 | | PSI-LHCI Supercomplex from Physcometrella patens | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhao, L, Yan, Q.J, Qin, X.C. | | Deposit date: | 2019-10-09 | | Release date: | 2021-02-10 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Antenna arrangement and energy-transfer pathways of PSI-LHCI from the moss Physcomitrella patens.
Cell Discov, 7, 2021
|
|
6L3F
 
 | |
6L8K
 
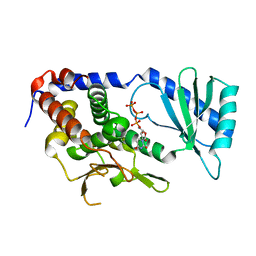 | | Structure of URT1 in complex with UTP | | Descriptor: | URIDINE 5'-TRIPHOSPHATE, UTP:RNA uridylyltransferase 1 | | Authors: | Lingru, Z. | | Deposit date: | 2019-11-06 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Crystal structure of Arabidopsis terminal uridylyl transferase URT1.
Biochem.Biophys.Res.Commun., 524, 2020
|
|
5ZGH
 
 | | Cryo-EM structure of the red algal PSI-LHCR | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Pi, X. | | Deposit date: | 2018-03-09 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Unique organization of photosystem I-light-harvesting supercomplex revealed by cryo-EM from a red alga
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5ZQ1
 
 | | Crystal structure of spRlmCD with U1939loop RNA at 3.10 angstrom | | Descriptor: | RNA (5'-R(*AP*AP*AP*(MUM)P*UP*CP*CP*U)-3'), S-ADENOSYL-L-HOMOCYSTEINE, Uncharacterized RNA methyltransferase SP_1029 | | Authors: | Yu, H.L, Jiang, Y.Y. | | Deposit date: | 2018-04-17 | | Release date: | 2018-10-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Unveiling the structural features that determine the dual methyltransferase activities of Streptococcus pneumoniae RlmCD
PLoS Pathog., 14, 2018
|
|
