1Q0Z
 
 | | Crystal structure of aclacinomycin methylesterase (RdmC) with bound product analogue, 10-decarboxymethylaclacinomycin A (DcmA) | | Descriptor: | 10-DECARBOXYMETHYLACLACINOMYCIN A (DCMAA), PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Jansson, A, Niemi, J, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-07-18 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of aclacinomycin methylesterase with bound product analogues: implications for anthracycline recognition and mechanism.
J.Biol.Chem., 278, 2003
|
|
1M5T
 
 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK | | Descriptor: | cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic and biochemical studies of DivK reveal novel features of an essential response regulator in Caulobacter crescentus
J.Biol.Chem., 277, 2002
|
|
1MB0
 
 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK AT PH 8.0 IN COMPLEX WITH MN2+ | | Descriptor: | MANGANESE (II) ION, cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-08-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and Biochemical Studies of DivK Reveal Novel Features of
an Essential Response Regulator in Caulobacter crescentus.
J.Biol.Chem., 277, 2002
|
|
1NAQ
 
 | | Crystal structure of CUTA1 from E.coli at 1.7 A resolution | | Descriptor: | MERCURIBENZOIC ACID, MERCURY (II) ION, Periplasmic divalent cation tolerance protein cutA | | Authors: | Calderone, V, Mangani, S, Benvenuti, M, Viezzoli, M.S, Banci, L, Bertini, I, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-11-28 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The evolutionarily conserved trimeric structure of CutA1 proteins suggests a role in signal transduction.
J.Biol.Chem., 278, 2003
|
|
2EW9
 
 | |
3BFJ
 
 | | Crystal structure analysis of 1,3-propanediol oxidoreductase | | Descriptor: | 1,3-propanediol oxidoreductase, FE (II) ION | | Authors: | Marcal, D, Enguita, F.J, Carrondo, M.A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-11-21 | | Release date: | 2008-11-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 1,3-propanediol dehydrogenase from Klebsiella pneumoniae: decameric quaternary structure and possible subunit cooperativity
J.Bacteriol., 191, 2009
|
|
1ZRZ
 
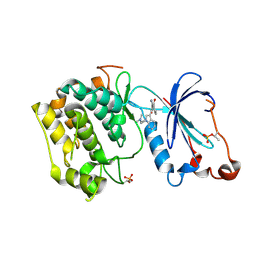 | | Crystal Structure of the Catalytic Domain of Atypical Protein Kinase C-iota | | Descriptor: | 3-{1-[3-(DIMETHYLAMINO)PROPYL]-1H-INDOL-3-YL}-4-(1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, Protein kinase C, iota | | Authors: | Messerschmidt, A, Macieira, S, Velarde, M, Baedeker, M, Benda, C, Jestel, A, Brandstetter, H, Neuefeind, T, Blaesse, M, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-05-23 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Atypical Protein Kinase C-iota Reveals Interaction Mode of Phosphorylation Site in Turn Motif
J.Mol.Biol., 352, 2005
|
|
1ZRU
 
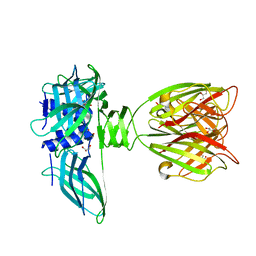 | | structure of the lactophage p2 receptor binding protein in complex with glycerol | | Descriptor: | GLYCEROL, lactophage p2 receptor binding protein | | Authors: | Spinelli, S, Tremblay, D.M, Tegoni, M, Blangy, S, Huyghe, C, Desmyter, A, Labrie, S, de Haard, H, Moineau, S, Cambillau, C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-05-22 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Receptor-binding protein of Lactococcus lactis phages: identification and characterization of the saccharide receptor-binding site.
J.Bacteriol., 188, 2006
|
|
2JCB
 
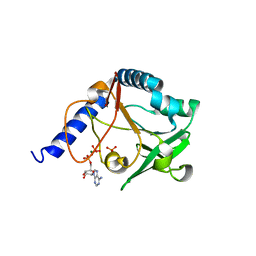 | | The crystal structure of 5-formyl-tetrahydrofolate cycloligase from Bacillus anthracis (BA4489) | | Descriptor: | 5-FORMYLTETRAHYDROFOLATE CYCLO-LIGASE FAMILY PROTEIN, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Meier, C, Carter, L.G, Winter, G, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-12-21 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of 5-Formyltetrahydrofolate Cyclo-Ligase from Bacillus Anthracis (Ba4489).
Acta Crystallogr.,Sect.F, 63, 2007
|
|
3A9E
 
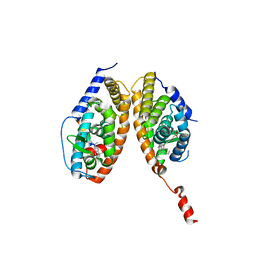 | | Crystal structure of a mixed agonist-bound RAR-alpha and antagonist-bound RXR-alpha heterodimer ligand binding domains | | Descriptor: | (2E,4E,6Z)-3-methyl-7-(5,5,8,8-tetramethyl-3-propoxy-5,6,7,8-tetrahydronaphthalen-2-yl)octa-2,4,6-trienoic acid, 13-mer (LXXLL motif) from Nuclear receptor coactivator 2, RETINOIC ACID, ... | | Authors: | Sato, Y, Duclaud, S, Peluso-Iltis, C, Poussin, P, Moras, D, Rochel, N, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2009-10-24 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Phantom Effect of the Rexinoid LG100754: structural and functional insights
Plos One, 5, 2010
|
|
1YUR
 
 | | Solution structure of apo-S100A13 (minimized mean structure) | | Descriptor: | S100 calcium-binding protein A13 | | Authors: | Arnesano, F, Banci, L, Bertini, I, Fantoni, A, Tenori, L, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-02-14 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Interplay between Calcium(II) and Copper(II) Binding to S100A13 Protein
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1YJE
 
 | | Crystal structure of the rNGFI-B ligand-binding domain | | Descriptor: | Orphan nuclear receptor NR4A1 | | Authors: | Flaig, R, Greschik, H, Peluso-Iltis, C, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-14 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the cell-specific activities of the NGFI-B and the Nurr1 ligand-binding domain.
J.Biol.Chem., 280, 2005
|
|
1YUT
 
 | | Solution structure of Calcium-S100A13 (minimized mean structure) | | Descriptor: | CALCIUM ION, S100 calcium-binding protein A13 | | Authors: | Arnesano, F, Banci, L, Bertini, I, Fantoni, A, Tenori, L, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-02-14 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Interplay between Calcium(II) and Copper(II) Binding to S100A13 Protein
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1YUS
 
 | | Solution structure of apo-S100A13 | | Descriptor: | S100 calcium binding protein A13 | | Authors: | Arnesano, F, Banci, L, Bertini, I, Fantoni, A, Tenori, L, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-02-14 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Interplay between Calcium(II) and Copper(II) Binding to S100A13 Protein
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1YUU
 
 | | Solution structure of Calcium-S100A13 | | Descriptor: | CALCIUM ION, S100 calcium-binding protein A13 | | Authors: | Arnesano, F, Banci, L, Bertini, I, Fantoni, A, Tenori, L, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-02-14 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Interplay between Calcium(II) and Copper(II) Binding to S100A13 Protein
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1XE3
 
 | | Crystal Structure of purine nucleoside phosphorylase DeoD from Bacillus anthracis | | Descriptor: | CHLORIDE ION, purine nucleoside phosphorylase | | Authors: | Grenha, R, Levdikov, V.M, Fogg, M, Blagova, E.V, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-09-09 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure of purine nucleoside phosphorylase (DeoD) from Bacillus anthracis.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1YLK
 
 | | Crystal Structure of Rv1284 from Mycobacterium tuberculosis in Complex with Thiocyanate | | Descriptor: | Hypothetical protein Rv1284/MT1322, THIOCYANATE ION, ZINC ION | | Authors: | Covarrubias, A.S, Larsson, A.M, Hogbom, M, Lindberg, J, Bergfors, T, Bjorkelid, C, Mowbray, S.L, Unge, T, Jones, T.A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-19 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of carbonic anhydrases from Mycobacterium tuberculosis.
J.Biol.Chem., 280, 2005
|
|
1YPF
 
 | | Crystal Structure of GuaC (BA5705) from Bacillus anthracis at 1.8 A Resolution | | Descriptor: | GMP reductase | | Authors: | Grenha, R, Levdikov, V.M, Blagova, E.V, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-31 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of GuaC (BA5705) from Bacillus anthracis at 1.8 A Resolution.
To be Published
|
|
1XA3
 
 | | Crystal structure of CaiB, a type III CoA transferase in carnitine metabolism | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Crotonobetainyl-CoA:carnitine CoA-transferase, SULFATE ION | | Authors: | Stenmark, P, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of CaiB, a Type-III CoA Transferase in Carnitine Metabolism
Biochemistry, 43, 2004
|
|
1YCM
 
 | | Solution Structure of matrix metalloproteinase 12 (MMP12) in the presence of N-Isobutyl-N-[4-methoxyphenylsulfonyl]glycyl hydroxamic acid (NNGH) | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Bertini, I, Calderone, V, Cosenza, M, Fragai, M, Lee, Y.M, Luchinat, C, Mangani, S, Terni, B, Turano, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-12-22 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformational variability of matrix metalloproteinases: Beyond a single 3D structure.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1Y3K
 
 | | Solution structure of the apo form of the fifth domain of Menkes protein | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Chasapis, C.T, Ciofi-Baffoni, S, Hadjiliadis, N, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-11-25 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An NMR study of the interaction between the human copper(I) chaperone and the second and fifth metal-binding domains of the Menkes protein
Febs J., 272, 2005
|
|
1XP3
 
 | | Crystal Structure of Endonuclease IV (BA4508) from Bacillus anthracis at 2.57A Resolution. | | Descriptor: | SULFATE ION, ZINC ION, endonuclease IV | | Authors: | Fogg, M.J, Levdikov, V.M, Blagova, E.V, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-10-08 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal Structure of Endonuclease IV (BA4508) from Bacillus anthracis at 2.57A Resolution.
To be Published
|
|
1Z2G
 
 | | Solution structure of apo, oxidized yeast Cox17 | | Descriptor: | Cytochrome c oxidase copper chaperone | | Authors: | Arnesano, F, Balatri, E, Banci, L, Bertini, I, Winge, D.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-03-08 | | Release date: | 2005-06-07 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Folding studies of Cox17 reveal an important interplay of cysteine oxidation and copper binding
STRUCTURE, 13, 2005
|
|
1Y3J
 
 | | Solution structure of the copper(I) form of the fifth domain of Menkes protein | | Descriptor: | COPPER (II) ION, Copper-transporting ATPase 1 | | Authors: | Banci, L, Chasapis, C.T, Ciofi-Baffoni, S, Hadjiliadis, N, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-11-25 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An NMR study of the interaction between the human copper(I) chaperone and the second and fifth metal-binding domains of the Menkes protein
Febs J., 272, 2005
|
|
1YM3
 
 | | Crystal Structure of carbonic anhydrase RV3588c from Mycobacterium tuberculosis | | Descriptor: | CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE), MAGNESIUM ION, ZINC ION | | Authors: | Covarrubias, A.S, Larsson, A.M, Hogbom, M, Lindberg, J, Bergfors, T, Bjorkelid, C, Mowbray, S.L, Unge, T, Jones, T.A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-20 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and function of carbonic anhydrases from Mycobacterium tuberculosis.
J.Biol.Chem., 280, 2005
|
|
