2DMJ
 
 | |
2D96
 
 | |
2D8K
 
 | |
2DM4
 
 | | Solution structure of the second fn3 domain of human sorLA/LR11 | | Descriptor: | Sortilin-related receptor | | Authors: | Nagashima, T, Kurosaki, C, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-20 | | Release date: | 2006-10-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second fn3 domain of human sorLA/LR11
To be Published
|
|
2E2W
 
 | |
2E32
 
 | | Structural basis for selection of glycosylated substrate by SCFFbs1 ubiquitin ligase | | Descriptor: | F-box only protein 2, S-phase kinase-associated protein 1A | | Authors: | Mizushima, T, Yoshida, Y, Kumanomidou, T, Hasegawa, Y, Yamane, T, Tanaka, K. | | Deposit date: | 2006-11-20 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Structural basis for the selection of glycosylated substrates by SCFFbs1 ubiquitin ligase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2EBU
 
 | |
2E33
 
 | | Structural basis for selection of glycosylated substrate by SCFFbs1 ubiquitin ligase | | Descriptor: | F-box only protein 2, Ribonuclease pancreatic, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Mizushima, T, Yoshida, Y, Kumanomidou, T, Hasegawa, Y, Yamane, T, Tanaka, K. | | Deposit date: | 2006-11-20 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the selection of glycosylated substrates by SCFFbs1 ubiquitin ligase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2ENZ
 
 | |
2EOD
 
 | |
2E31
 
 | | Structural basis for selection of glycosylated substrate by SCFFbs1 ubiquitin ligase | | Descriptor: | F-box only protein 2, S-phase kinase-associated protein 1A | | Authors: | Mizushima, T, Yoshida, Y, Kumanomidou, T, Hasegawa, Y, Yamane, T, Tanaka, K. | | Deposit date: | 2006-11-20 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the selection of glycosylated substrates by SCFFbs1 ubiquitin ligase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2DC0
 
 | | Crystal structure of amidase | | Descriptor: | probable amidase | | Authors: | Ohshima, T, Sakuraba, H, Ebihara, A, Kanagawa, M, Nakagawa, N, Kuroishi, C, Satoh, S, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-17 | | Release date: | 2007-01-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of amidase
To be Published
|
|
6LHR
 
 | |
6LII
 
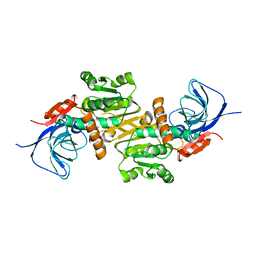 | | A quinone oxidoreductase | | Descriptor: | Synaptic vesicle membrane protein VAT-1 homolog | | Authors: | Hakoshima, T, Kim, S.-Y, Mori, T. | | Deposit date: | 2019-12-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into vesicle amine transport-1 (VAT-1) as a member of the NADPH-dependent quinone oxidoreductase family.
Sci Rep, 11, 2021
|
|
8IC8
 
 | | Exo-alpha-D-arabinofuranosidase from Microbacterium arabinogalactanolyticum | | Descriptor: | Exo-alpha-D-arabinofuranosidase, PHOSPHATE ION | | Authors: | Kashima, T, Arakawa, T, Yamada, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
8HPK
 
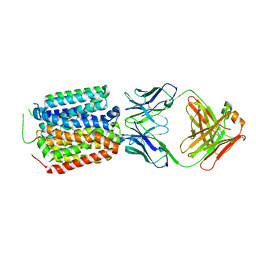 | | Crystal structure of the bacterial oxalate transporter OxlT in an oxalate-bound occluded form | | Descriptor: | Fab fragment Heavy chein, Fab fragment Light chain, OXALATE ION, ... | | Authors: | Shimamura, T, Hirai, T, Yamashita, A. | | Deposit date: | 2022-12-12 | | Release date: | 2023-02-15 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and mechanism of oxalate transporter OxlT in an oxalate-degrading bacterium in the gut microbiota.
Nat Commun, 14, 2023
|
|
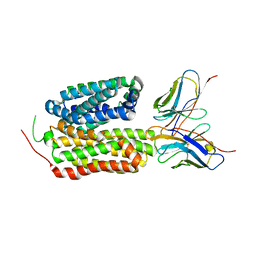
8HPJ
 
 | |
4QBJ
 
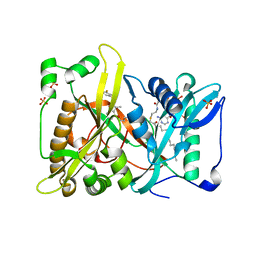 | | Crystal structure of N-myristoyl transferase from Aspergillus fumigatus complexed with a synthetic inhibitor | | Descriptor: | 3-[[3-methyl-2-[[2,3,4-tris(fluoranyl)phenoxy]methyl]-1-benzofuran-4-yl]oxy]-N-(pyridin-3-ylmethyl)propan-1-amine, Glycylpeptide N-tetradecanoyltransferase, S-(2-OXO)PENTADECYLCOA, ... | | Authors: | Suzuki, M, Shimada, T. | | Deposit date: | 2014-05-08 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of N-myristoyltransferase from Aspergillus fumigatus
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7WDU
 
 | | 6-sulfo-beta-D-N-acetylglucosaminidase from Bifidobacterium bifidum in complex with PUGNAc-6S | | Descriptor: | Beta-N-acetylhexosaminidase, CALCIUM ION, [[(3R,4R,5S,6R)-3-acetamido-4,5-bis(oxidanyl)-6-(sulfooxymethyl)oxan-2-ylidene]amino] N-phenylcarbamate | | Authors: | Kashima, T, Yamada, C, Fushinobu, S, Katoh, T, Katayama, T. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | A bacterial sulfoglycosidase highlights mucin O-glycan breakdown in the gut ecosystem.
Nat.Chem.Biol., 19, 2023
|
|
2RQJ
 
 | | Quadruplex structure of an RNA aptamer against bovine prion protein | | Descriptor: | RNA (5'-R(*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*A)-3') | | Authors: | Katahira, M, Mashima, T, Matsugami, A, Nishikawa, F, Nishikawa, S. | | Deposit date: | 2009-07-18 | | Release date: | 2009-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Unique quadruplex structure and interaction of an RNA aptamer against bovine prion protein
Nucleic Acids Res., 37, 2009
|
|
5T1F
 
 | | Crystal structure of Phaeospaeria nodrum fructosyl peptide oxidase mutant Asn56Ala | | Descriptor: | ACETIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Uncharacterized protein | | Authors: | Yoshida, H, Shimasaki, T, Kamitori, S, Sode, K. | | Deposit date: | 2016-08-19 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction
Sci Rep, 7, 2017
|
|
5T1E
 
 | | Crystal structure of Phaeospaeria nodrum fructosyl peptide oxidase | | Descriptor: | ACETIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Uncharacterized protein | | Authors: | Yoshida, H, Shimasaki, T, Kamitori, S, Sode, K. | | Deposit date: | 2016-08-19 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction
Sci Rep, 7, 2017
|
|
5YQ5
 
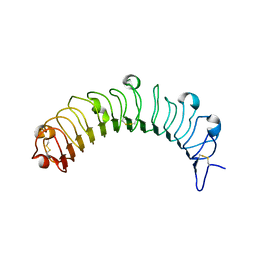 | | Crystal structure of human osteomodulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Osteomodulin, PHOSPHATE ION | | Authors: | Caaveiro, J.M.M, Tashima, T, Tsumoto, K. | | Deposit date: | 2017-11-05 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Molecular basis for governing the morphology of type-I collagen fibrils by Osteomodulin.
Commun Biol, 1, 2018
|
|
2ZIY
 
 | | Crystal structure of squid rhodopsin | | Descriptor: | PALMITIC ACID, RETINAL, Rhodopsin | | Authors: | Miyano, M, Shimamura, T. | | Deposit date: | 2008-02-27 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of squid rhodopsin with intracellularly extended cytoplasmic region
J.Biol.Chem., 283, 2008
|
|
2Z41
 
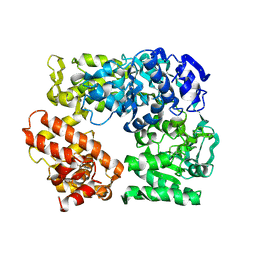 | | Crystal Structure Analysis of the Ski2-type RNA helicase | | Descriptor: | MAGNESIUM ION, putative ski2-type helicase | | Authors: | Nakashima, T, Zhang, X, Kakuta, Y, Yao, M, Tanaka, I, Kimura, M. | | Deposit date: | 2007-06-12 | | Release date: | 2008-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Crystal structure of an archaeal Ski2p-like protein from Pyrococcus horikoshii OT3
Protein Sci., 17, 2008
|
|
