4U40
 
 | |
4U42
 
 | | MAP4K4 T181E Mutant Bound to inhibitor compound 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[(3S)-3-(4-methyl-1H-pyrazol-3-yl)piperidin-1-yl]pyrido[3,2-d]pyrimidin-4-amine, MAGNESIUM ION, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2016-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural Plasticity and Kinase Activation in a Cohort of MAP4K4 Structures
to be published
|
|
4U3Y
 
 | |
4U41
 
 | | MAP4K4 Bound to inhibitor compound 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[(3S)-3-(4-methyl-1H-pyrazol-3-yl)piperidin-1-yl]pyrido[3,2-d]pyrimidin-4-amine, MAGNESIUM ION, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2016-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Plasticity and Kinase Activation in a Cohort of MAP4K4 Structures
to be published
|
|
4U45
 
 | | MAP4K4 in complex with inhibitor (compound 25) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(1H-pyrazol-4-yl)-N-(pyridin-4-yl)pyrrolo[2,1-f][1,2,4]triazin-4-amine, MAGNESIUM ION, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Fragment-based identification and optimization of a class of potent pyrrolo[2,1-f][1,2,4]triazine MAP4K4 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5HVY
 
 | | CDK8/CYCC IN COMPLEX WITH COMPOUND 20 | | Descriptor: | CHLORIDE ION, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Kiefer, J.R, Schneider, E.V, Maskos, K, Bergeron, P, Koehler, M. | | Deposit date: | 2016-01-28 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Design and Development of a Series of Potent and Selective Type II Inhibitors of CDK8.
Acs Med.Chem.Lett., 7, 2016
|
|
6UYC
 
 | | Crystal structure of TEAD2 bound to Compound 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-{5-[(E)-2-(4,4-difluorocyclohexyl)ethenyl]-6-methoxypyridin-3-yl}methanesulfonamide, Transcriptional enhancer factor TEF-4 | | Authors: | Noland, C.L, Holden, J.K, Crawford, J.J, Zbieg, J.R, Cunningham, C.N. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.658 Å) | | Cite: | Small Molecule Dysregulation of TEAD Lipidation Induces a Dominant-Negative Inhibition of Hippo Pathway Signaling.
Cell Rep, 31, 2020
|
|
6UYB
 
 | | Crystal structure of TEAD2 bound to Compound 1 | | Descriptor: | (3R,4R)-1-{3-[(E)-2-(4-chlorophenyl)ethenyl]-4-methoxy-5-methylphenyl}-3,4-dihydroxypyrrolidin-2-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Noland, C.L, Holden, J.K, Crawford, J.J, Zbieg, J.R, Cunningham, C.N. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Small Molecule Dysregulation of TEAD Lipidation Induces a Dominant-Negative Inhibition of Hippo Pathway Signaling.
Cell Rep, 31, 2020
|
|
4ZK5
 
 | | MAP4K4 in complex with inhibitor GNE-495 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 8-amino-N-[1-(cyclopropylcarbonyl)azetidin-3-yl]-2-(3-fluorophenyl)-1,7-naphthyridine-5-carboxamide, MAGNESIUM ION, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2015-04-29 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure-Based Design of GNE-495, a Potent and Selective MAP4K4 Inhibitor with Efficacy in Retinal Angiogenesis.
Acs Med.Chem.Lett., 6, 2015
|
|
6R2G
 
 | | Crystal structure of a single-chain protein mimetic of the gp41 NHR trimer in complex with the synthetic CHR peptide C34 | | Descriptor: | Envelope glycoprotein gp160, PHOSPHATE ION, Single-chain protein mimetics of the N-terminal heptad-repeat region of gp41 | | Authors: | Camara-Artigas, A, Conejero-Lara, F, Jurado, S, Cano-Munoz, M, Morel, B. | | Deposit date: | 2019-03-17 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Thermodynamic Analysis of HIV-1 Fusion Inhibition Using Small gp41 Mimetic Proteins.
J.Mol.Biol., 431, 2019
|
|
2YHA
 
 | |
1LBM
 
 | |
2YKO
 
 | |
2YHB
 
 | |
2YKP
 
 | |
2YKQ
 
 | |
4C1B
 
 | |
4C1A
 
 | |
4QYE
 
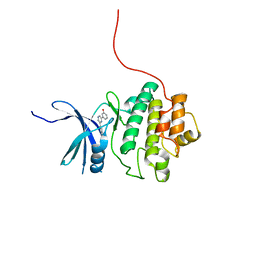 | | CHK1 kinase domain in complex with diarylpyrazine compound 1 | | Descriptor: | 4-[6-(3-hydroxyphenyl)pyrazin-2-yl]benzoic acid, Serine/threonine-protein kinase Chk1 | | Authors: | Appleton, B, Wu, P. | | Deposit date: | 2014-07-24 | | Release date: | 2014-12-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of the 1,7-diazacarbazole class of inhibitors of checkpoint kinase 1.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
7BL4
 
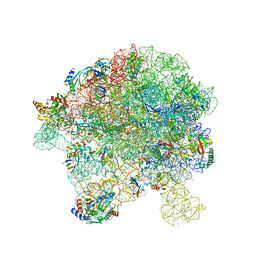 | | in vitro reconstituted 50S-ObgE-GMPPNP-RsfS particle | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Hilal, T, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
7BL3
 
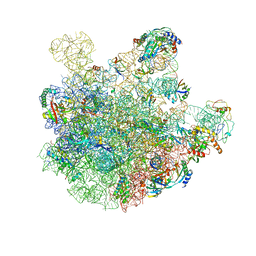 | | pre-50S-ObgE particle state 2 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Hilal, T, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
6T6I
 
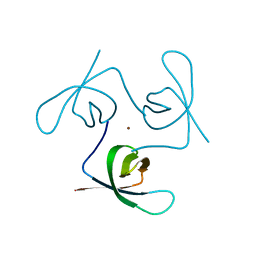 | |
6T6E
 
 | |
6T6J
 
 | |
5AOX
 
 | |
