4Z8Z
 
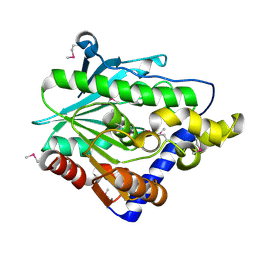 | | Crystal structure of the hypothetical protein from Ruminiclostridium thermocellum ATCC 27405 | | Descriptor: | Uncharacterized protein | | Authors: | Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Endres, M, Joachimiak, J, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-04-09 | | Release date: | 2015-05-06 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the hypothetical protein from Ruminiclostridium thermocellum ATCC 27405
To Be Published
|
|
4ZPJ
 
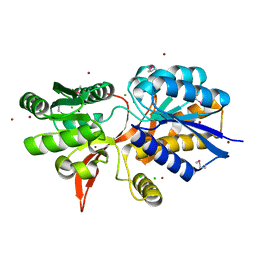 | | ABC transporter substrate-binding protein from Sphaerobacter thermophilus | | Descriptor: | CHLORIDE ION, Extracellular ligand-binding receptor, ZINC ION | | Authors: | OSIPIUK, J, Holowicki, J, Clancy, S, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-07 | | Release date: | 2015-05-20 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | ABC transporter substrate-binding protein from Sphaerobacter thermophilus.
to be published
|
|
5DYF
 
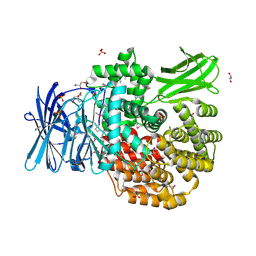 | | The crystal structure of Aminopeptidase N in complex with N-benzyl-1,2-diaminoethylphosphonic acid | | Descriptor: | Aminopeptidase N, GLYCEROL, IMIDAZOLE, ... | | Authors: | Nocek, B, Joachimiak, A, Vassiliou, S, Berlicki, L, Mucha, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-09-24 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | The crystal structure of Aminopeptidase N in complex with N-benzyl-1,2-diaminoethylphosphonic acid
To Be Published
|
|
5TGN
 
 | | Crystal structure of protein Sthe_2403 from Sphaerobacter thermophilus | | Descriptor: | CHLORIDE ION, GLYCEROL, Uncharacterized protein | | Authors: | Michalska, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-28 | | Release date: | 2016-10-26 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of protein Sthe_2403 from Sphaerobacter thermophilus
To Be Published
|
|
5TJJ
 
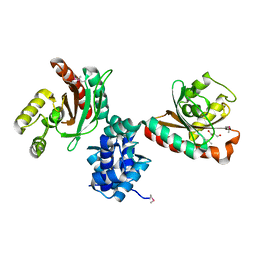 | | Crystal structure of IcIR transcriptional regulator from Alicyclobacillus acidocaldarius | | Descriptor: | GLYCEROL, Transcriptional regulator, IclR family | | Authors: | Michalska, K, Mack, J.C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-10-04 | | Release date: | 2016-10-26 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of IcIR transcriptional regulator from Alicyclobacillus acidocaldarius
To Be Published
|
|
5TTX
 
 | |
5UVE
 
 | | Crystal Structure of the ABC Transporter Substrate-binding protein BAB1_0226 from Brucella abortus | | Descriptor: | CALCIUM ION, GLYCEROL, Substrate-binding region of ABC-type glycine betaine transport system | | Authors: | Kim, Y, Chhor, G, Endres, M, Hero, J, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-02-20 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Beta-barrel-like Protein of Unknown Function
To Be Published
|
|
5T2X
 
 | | Crystal structure of Uncharacterised protein lpg1670 | | Descriptor: | Uncharacterized protein LPG1670 | | Authors: | Chang, C, Xu, X, Cui, H, SAVCHENKO, A, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-08-24 | | Release date: | 2016-09-28 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Crystal structure of Uncharacterised protein lpg1670
To Be Published
|
|
4DWJ
 
 | | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in complex with Thymidine Monophosphate | | Descriptor: | THYMIDINE-5'-PHOSPHATE, Thymidylate kinase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-02-24 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in complex with Thymidine Monophosphate
To be Published
|
|
4DIM
 
 | |
4EVX
 
 | | Crystal structure of putative phage endolysin from S. enterica | | Descriptor: | Putative phage endolysin | | Authors: | Michalska, K, Li, H, Jedrzejczak, R, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Adkins, J.N, Joachimiak, A, Program for the Characterization of Secreted Effector Proteins (PCSEP), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-26 | | Release date: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative phage endolysin from S. enterica
To be Published
|
|
4ESY
 
 | | Crystal Structure of the CBS Domain of CBS Domain Containing Membrane Protein from Sphaerobacter thermophilus | | Descriptor: | 1,2-ETHANEDIOL, CBS domain containing membrane protein, CHLORIDE ION | | Authors: | Kim, Y, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-23 | | Release date: | 2012-09-05 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Crystal Structure of the CBS Domain of CBS Domain Containing Membrane Protein from Sphaerobacter thermophilus
To be Published
|
|
4FB7
 
 | | The apo form of idole-3-glycerol phosphate synthase (TrpC) form Mycobacterium tuberculosis | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Indole-3-glycerol phosphate synthase | | Authors: | Michalska, K, Chhor, G, Jedrzejczak, R, Terwilliger, T.C, Rubin, E.J, Guinn, K, Baker, D, Ioerger, T.R, Sacchettini, J.C, Joachimiak, A, Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-22 | | Release date: | 2012-06-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The apo form of idole-3-glycerol phosphate synthase (TrpC) form Mycobacterium tuberculosis
To be Published
|
|
4G6X
 
 | | Crystal structure of glyoxalase/bleomycin resistance protein from Catenulispora acidiphila. | | Descriptor: | GLYCEROL, Glyoxalase/bleomycin resistance protein/dioxygenase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-07-19 | | Release date: | 2012-08-15 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of glyoxalase/bleomycin resistance protein from Catenulispora acidiphila.
To be Published
|
|
1VR4
 
 | | Crystal Structure of MCSG TArget APC22750 from Bacillus cereus | | Descriptor: | hypothetical protein APC22750 | | Authors: | Yang, X, Brunzelle, J.S, McNamara, L.K, Minasov, G, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-01-31 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of MCSG TArget APC22750 from Bacillus cereus
To be Published
|
|
5TFQ
 
 | | Crystal structure of a representative of class A beta-lactamase from Bacteroides cellulosilyticus DSM 14838 | | Descriptor: | Beta-lactamase, GLYCEROL, PHOSPHATE ION | | Authors: | Nocek, B, Hatzos-Skintges, C, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-26 | | Release date: | 2016-11-02 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Crystal structure of a representative of class A beta-lactamase from Bacteroides cellulosilyticus DSM 14838
To Be Published
|
|
5TGF
 
 | | Crystal structure of putative beta-lactamase from Bacteroides dorei DSM 17855 | | Descriptor: | CALCIUM ION, GLYCEROL, Uncharacterized protein | | Authors: | Nocek, B, Hatzos-Skintges, C, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-27 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of a beta-lactamase from Bacteroides dorei DSM 17855
To Be Published
|
|
5W27
 
 | | Crystal structure of TnmS3 in complex with tiancimycin (TNM B) | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, methyl (2E)-3-[(1aS,11S,11aS,14Z,18R)-3,18-dihydroxy-4,9-dioxo-4,9,10,11-tetrahydro-11aH-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinolin-11a-yl]but-2-enoate | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, SHen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-06-05 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of TnmS3 in complex with tiancimycin (TNM B)
To Be Published
|
|
1TT4
 
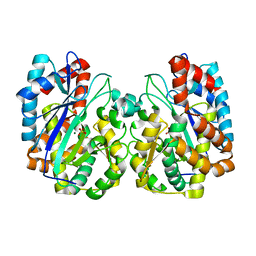 | | Structure of NP459575, a predicted glutathione synthase from Salmonella typhimurium | | Descriptor: | MAGNESIUM ION, SULFATE ION, putative cytoplasmic protein | | Authors: | Miller, D.J, Shuvalova, L, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-21 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structure of NP459575, a predicted glutathione synthase from Salmonella typhimurium
To be Published
|
|
1U84
 
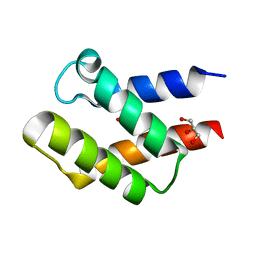 | | Crystal Structure of APC36109 from Bacillus stearothermophilus | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Hypothetical protein | | Authors: | Kim, Y, Zhou, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-04 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of APC36109 from Bacillus stearothermophilus
To be Published
|
|
1W8I
 
 | | The Structure of gene product af1683 from Archaeoglobus fulgidus. | | Descriptor: | PUTATIVE VAPC RIBONUCLEASE AF_1683 | | Authors: | Midwest Center for Structural Genomics (MCSG), Cuff, M.E, Zhang, R, Ginell, S.L, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A. | | Deposit date: | 2004-09-22 | | Release date: | 2004-11-16 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of Gene Product Af1683 from Archaeoglobus Fulgidus
To be Published
|
|
2AH5
 
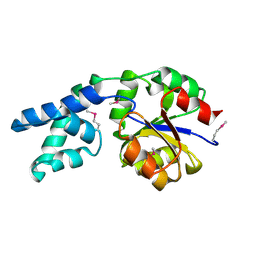 | | Hydrolase, haloacid dehalogenase-like family protein SP0104 from Streptococcus pneumoniae | | Descriptor: | COG0546: Predicted phosphatases | | Authors: | Binkowski, T.A, Zhou, M, Abdullah, J, Collart, F, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-07-27 | | Release date: | 2005-09-27 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Hydrolase, haloacid dehalogenase-like family protein SP0104 from Streptococcus pneumoniae
To be Published
|
|
4MK6
 
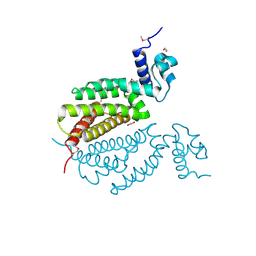 | | Crystal Structure of Probable Dihydroxyacetone Kinase Regulator DHSK_reg from Listeria monocytogenes EGD-e | | Descriptor: | 1,2-ETHANEDIOL, Probable Dihydroxyacetone Kinase Regulator DHSK_reg | | Authors: | Kim, Y, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Probable Dihydroxyacetone Kinase Regulator DHSK_reg from Listeria monocytogenes EGD-e
To be Published
|
|
4MO9
 
 | | Crystal Structure of TroA-like Periplasmic Binding Protein FepB from Veillonella parvula | | Descriptor: | GLYCEROL, Periplasmic binding protein, trimethylamine oxide | | Authors: | Kim, Y, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-11 | | Release date: | 2013-09-25 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | Crystal Structure of TroA-like Periplasmic Binding Protein FepB from Veillonella parvula
To be Published
|
|
1XA0
 
 | | Crystal Structure of MCSG Target APC35536 from Bacillus stearothermophilus | | Descriptor: | CHLORIDE ION, Putative NADPH Dependent oxidoreductases, SULFATE ION | | Authors: | Brunzelle, J.S, Sommerhalter, M, Minasov, G, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-24 | | Release date: | 2004-10-05 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of MCSG Target APC35536 from Bacillus stearothermophilus
To be Published
|
|
