5XZG
 
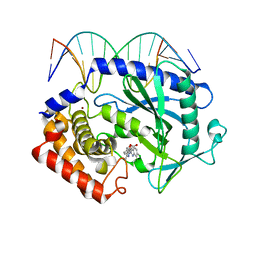 | | Mouse cGAS bound to the inhibitor RU521 | | Descriptor: | 2-(4,5-dichloro-1H-benzimidazol-2-yl)-5-methyl-4-[(1R)-3-oxo-1,3-dihydro-2-benzofuran-1-yl]-1,2-dihydro-3H-pyrazol-3-one, Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*G)-3'), ... | | Authors: | Vincent, J, Adura, C, Gao, P, Luz, A, Lama, L, Asano, Y, Okamoto, R, Imaeda, T, Aida, J, Rothamel, K, Gogakos, T, Steinberg, J, Reasoner, S, Aso, K, Tuschl, T, Patel, D.J, Glickman, J.F, Ascano, M. | | Deposit date: | 2017-07-12 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice.
Nat Commun, 8, 2017
|
|
5XZB
 
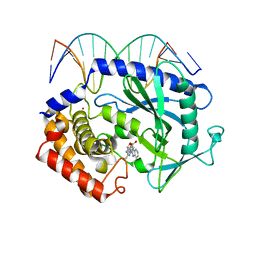 | | Mouse cGAS bound to the inhibitor RU365 | | Descriptor: | (3R)-3-[1-(1H-benzimidazol-2-yl)-5-hydroxy-3-methyl-1H-pyrazol-4-yl]-2-benzofuran-1(3H)-one, Cyclic GMP-AMP synthase, DNA (5'-D(*AP*AP*AP*TP*TP*GP*CP*CP*GP*AP*AP*GP*AP*CP*G)-3'), ... | | Authors: | Vincent, J, Adura, C, Gao, P, Luz, A, Lama, L, Asano, Y, Okamoto, R, Imaeda, T, Aida, J, Rothamel, K, Gogakos, T, Steinberg, J, Reasoner, S, Aso, K, Tuschl, T, Patel, D.J, Glickman, J.F, Ascano, M. | | Deposit date: | 2017-07-12 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice.
Nat Commun, 8, 2017
|
|
5X6D
 
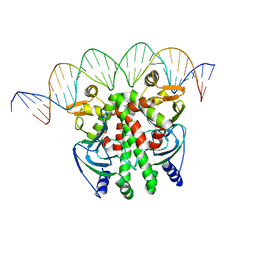 | | Crystal structure of PrfA-DNA binary complex | | Descriptor: | DNA (28-MER), DNA (29-MER), Listeriolysin positive regulatory factor A | | Authors: | Wang, Y, Feng, H, Zhu, Y.L, Gao, P. | | Deposit date: | 2017-02-21 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structural insights into glutathione-mediated activation of the master regulator PrfA in Listeria monocytogenes
Protein Cell, 8, 2017
|
|
5X6E
 
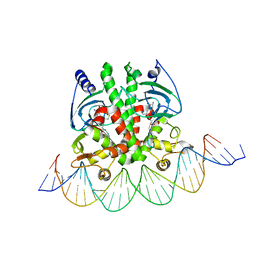 | | Crystal structure of PrfA-DNA binary complex | | Descriptor: | DNA (28-MER), DNA (29-MER), GLUTATHIONE, ... | | Authors: | Wang, Y, Feng, H, Zhu, Y.L, Gao, P. | | Deposit date: | 2017-02-21 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural insights into glutathione-mediated activation of the master regulator PrfA in Listeria monocytogenes
Protein Cell, 8, 2017
|
|
6WOJ
 
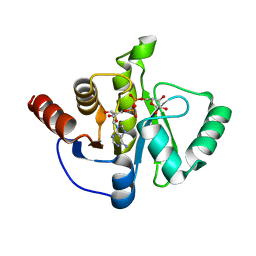 | | Structure of the SARS-CoV-2 macrodomain (NSP3) in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Gao, F.P, Fehr, A.R. | | Deposit date: | 2020-04-24 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The SARS-CoV-2 Conserved Macrodomain Is a Mono-ADP-Ribosylhydrolase.
J.Virol., 95, 2021
|
|
2RVH
 
 | | NMR structure of eIF1 | | Descriptor: | Eukaryotic translation initiation factor eIF-1 | | Authors: | Nagata, T, Obayashi, E, Asano, K. | | Deposit date: | 2015-10-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Landscape of the Ribosome Pre-initiation Complex during mRNA Scanning: Structural Role for eIF3c and Its Control by eIF5
Cell Rep, 18, 2017
|
|
9BK3
 
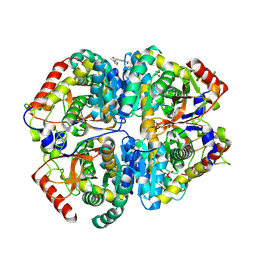 | | Crystal structure of Lactate dehydrogenase in complex with 4-((4-(1-methyl-1H-imidazole-2-carbonyl)phenyl)amino)-4-oxo-2-(4-(trifluoromethyl)phenyl)butanoic acid (R-enantiomer, orthorhombic P form) | | Descriptor: | (2R)-4-[4-(1-methyl-1H-imidazole-2-carbonyl)anilino]-4-oxo-2-[4-(trifluoromethyl)phenyl]butanoic acid, CHLORIDE ION, L-lactate dehydrogenase A chain, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Sharma, H. | | Deposit date: | 2024-04-26 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and biological characterization of an orally bioavailable lactate dehydrogenase-A inhibitor against pancreatic cancer.
Eur.J.Med.Chem., 275, 2024
|
|
9BK2
 
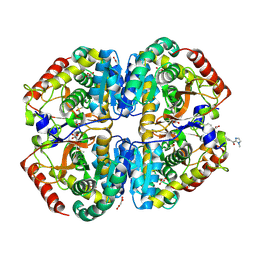 | | Crystal structure of Lactate dehydrogenase in complex with 4-((4-(1-methyl-1H-imidazole-2-carbonyl)phenyl)amino)-4-oxo-2-(4-(trifluoromethyl)phenyl)butanoic acid (S-enantiomer, monoclinic P form) | | Descriptor: | (2S)-4-[4-(1-methyl-1H-imidazole-2-carbonyl)anilino]-4-oxo-2-[4-(trifluoromethyl)phenyl]butanoic acid, DIMETHYL SULFOXIDE, L-lactate dehydrogenase A chain, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Sharma, H. | | Deposit date: | 2024-04-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis and biological characterization of an orally bioavailable lactate dehydrogenase-A inhibitor against pancreatic cancer.
Eur.J.Med.Chem., 275, 2024
|
|
7BQ5
 
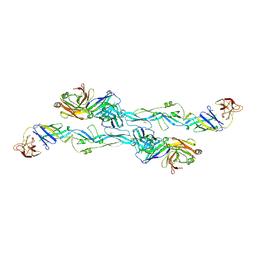 | | ZIKV sE bound to mAb Z6 | | Descriptor: | Z6 Light Chain, Z6 heavy chain, envelope protein | | Authors: | Dai, L, Qi, J, Gao, G.F. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Protective Zika vaccines engineered to eliminate enhancement of dengue infection via immunodominance switch.
Nat.Immunol., 22, 2021
|
|
7BPK
 
 | | Zika virus envelope protein mutant bound to mAb | | Descriptor: | Envelope protein, IG c307_light_IGLV1-51_IGLJ2, Z3L1 Heavy chain | | Authors: | Dai, L, Qi, J, Gao, G.F. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protective Zika vaccines engineered to eliminate enhancement of dengue infection via immunodominance switch.
Nat.Immunol., 22, 2021
|
|
9EQG
 
 | |
6DMQ
 
 | |
6DMM
 
 | | Crystal structure of the G23A mutant of human alpha defensin HNP4. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Neutrophil defensin 4, SULFATE ION | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2018-06-05 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Systematic mutational analysis of human neutrophil alpha-defensin HNP4.
Biochim Biophys Acta Biomembr, 1861, 2019
|
|
3VE5
 
 | |
3VDU
 
 | |
6NTY
 
 | | 2.1 A resolution structure of the Musashi-2 (Msi2) RNA recognition motif 1 (RRM1) domain | | Descriptor: | PHOSPHATE ION, RNA-binding protein Musashi homolog 2 | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Lan, L, Xiaoqing, W, Cooper, A, Gao, F.P, Xu, L. | | Deposit date: | 2019-01-30 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal and solution structures of human oncoprotein Musashi-2 N-terminal RNA recognition motif 1.
Proteins, 88, 2020
|
|
8TGB
 
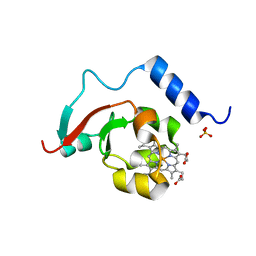 | | Crystal structure of root lateral formation protein (RLF) b5-domain from Oryza sativa | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, root lateral formation protein (RLF) | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Benson, D.R. | | Deposit date: | 2023-07-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The N-terminal intrinsically disordered region of Ncb5or docks with the cytochrome b 5 core to form a helical motif that is of ancient origin.
Proteins, 92, 2024
|
|
3VDP
 
 | |
8K4F
 
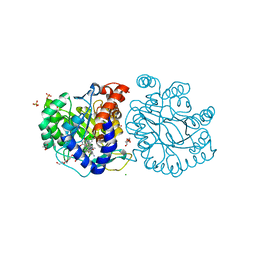 | | DHODH in complex with compound A0 | | Descriptor: | 5-cyclopropyl-2-[1-[(2-fluorophenyl)methyl]pyrazolo[3,4-b]pyridin-3-yl]pyrimidin-4-amine, 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, ACETATE ION, ... | | Authors: | Jian, L, Sun, Q. | | Deposit date: | 2023-07-18 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery and Optimization of Novel h DHODH Inhibitors for the Treatment of Inflammatory Bowel Disease.
J.Med.Chem., 66, 2023
|
|
8HR7
 
 | | Structure of RdrA-RdrB complex | | Descriptor: | Adenosine deaminase, Archaeal ATPase | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HR8
 
 | | Structure of heptameric RdrA ring | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HRA
 
 | | Structure of heptameric RdrA ring in RNA-loading state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase, RNA (5'-R(P*GP*UP*CP*CP*AP*GP*CP*GP*UP*CP*AP*UP*CP*GP*CP*UP*GP*GP*AP*C)-3') | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HRC
 
 | | Structure of dodecameric RdrB cage | | Descriptor: | Adenosine deaminase | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HRB
 
 | | Structure of tetradecameric RdrA ring in RNA-loading state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase, RNA (5'-R(*GP*UP*CP*CP*AP*GP*CP*GP*UP*CP*AP*UP*CP*GP*CP*UP*GP*GP*AP*C)-3') | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HR9
 
 | | Structure of tetradecameric RdrA ring | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
