4U3E
 
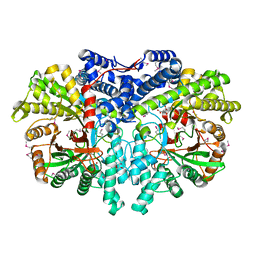 | | Anaerobic ribonucleotide reductase | | Descriptor: | ACETATE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2014-07-20 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The class III ribonucleotide reductase from Neisseria bacilliformis can utilize thioredoxin as a reductant.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
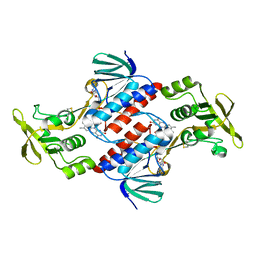
3CTY
 
 | |
3GJB
 
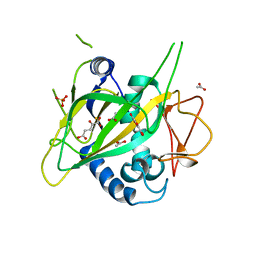 | | CytC3 with Fe(II) and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, CytC3, ... | | Authors: | Wong, C, Drennan, C.L. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of an open active site conformation of nonheme iron halogenase CytC3
J.Am.Chem.Soc., 131, 2009
|
|
3GJA
 
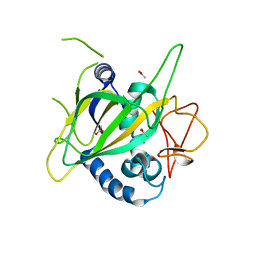 | | CytC3 | | Descriptor: | ACETATE ION, CytC3 | | Authors: | Wong, C, Drennan, C.L. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of an open active site conformation of nonheme iron halogenase CytC3
J.Am.Chem.Soc., 131, 2009
|
|
1Q5V
 
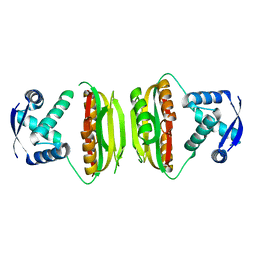 | | Apo-NikR | | Descriptor: | Nickel responsive regulator | | Authors: | Schreiter, E.R, Sintchak, M.D, Guo, Y, Chivers, P.T, Sauer, R.T, Drennan, C.L. | | Deposit date: | 2003-08-11 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Nickel-Responsive Transcription Factor NikR
Nat.Struct.Biol., 10, 2003
|
|
1Q5Y
 
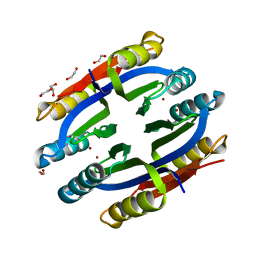 | | Nickel-Bound C-terminal Regulatory Domain of NikR | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Nickel responsive regulator | | Authors: | Schreiter, E.R, Sintchak, M.D, Guo, Y, Chivers, P.T, Sauer, R.T, Drennan, C.L. | | Deposit date: | 2003-08-11 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the Nickel-Responsive Transcription Factor NikR
Nat.Struct.Biol., 10, 2003
|
|
2Z8Y
 
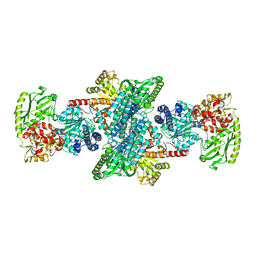 | | Xenon-bound structure of bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase(CODH/ACS) from Moorella thermoacetica | | Descriptor: | COPPER (I) ION, Carbon monoxide dehydrogenase/acetyl CoA synthase subunit alpha, Carbon monoxide dehydrogenase/acetyl CoA synthase subunit beta, ... | | Authors: | Doukov, T.I, Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2007-09-12 | | Release date: | 2008-03-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Xenon in and at the End of the Tunnel of Bifunctional Carbon Monoxide Dehydrogenase/Acetyl-CoA Synthase
Biochemistry, 47, 2008
|
|
3BMZ
 
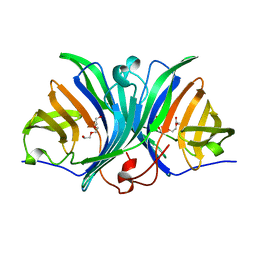 | | Violacein biosynthetic enzyme VioE | | Descriptor: | Putative uncharacterized protein, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Ryan, K.S, Drennan, C.L. | | Deposit date: | 2007-12-13 | | Release date: | 2008-01-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | The Violacein Biosynthetic Enzyme VioE Shares a Fold with Lipoprotein Transporter Proteins
J.Biol.Chem., 283, 2008
|
|
3BKT
 
 | |
3CB8
 
 | |
6OWR
 
 | | NMR solution structure of YfiD | | Descriptor: | Autonomous glycyl radical cofactor | | Authors: | Bowman, S.E.J, Drennan, C.L. | | Deposit date: | 2019-05-10 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and biochemical characterization of a spare part protein that restores activity to an oxygen-damaged glycyl radical enzyme.
J.Biol.Inorg.Chem., 24, 2019
|
|
6ONS
 
 | | Crystal structure of Desulfovibrio vulgaris carbon monoxide dehydrogenase with the D-cluster ligating cysteines mutated to alanines, coexpressed with CooC, as-isolated | | Descriptor: | Carbon monoxide dehydrogenase, FE(4)-NI(1)-S(4) CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Cohen, S.E, Wittenborn, E.C, Drennan, C.L. | | Deposit date: | 2019-04-22 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.485 Å) | | Cite: | Structural insight into metallocofactor maturation in carbon monoxide dehydrogenase.
J.Biol.Chem., 294, 2019
|
|
6ONC
 
 | | Crystal structure of Desulfovibrio vulgaris carbon monoxide dehydrogenase produced without CooC, as-isolated | | Descriptor: | CHLORIDE ION, Carbon monoxide dehydrogenase, FE (III) ION, ... | | Authors: | Wittenborn, E.C, Cohen, S.E, Drennan, C.L. | | Deposit date: | 2019-04-21 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insight into metallocofactor maturation in carbon monoxide dehydrogenase.
J.Biol.Chem., 294, 2019
|
|
7MDI
 
 | | Structure of the Neisseria gonorrhoeae ribonucleotide reductase in the inactive state | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Levitz, T.S, Drennan, C.L, Brignole, E.J. | | Deposit date: | 2021-04-05 | | Release date: | 2022-01-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Effects of chameleon dispense-to-plunge speed on particle concentration, complex formation, and final resolution: A case study using the Neisseria gonorrhoeae ribonucleotide reductase inactive complex.
J.Struct.Biol., 214, 2021
|
|
7LHR
 
 | |
7LHU
 
 | |
7LHS
 
 | |
2OAM
 
 | |
2OAL
 
 | | RebH with bound FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Tryptophan halogenase | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2006-12-16 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chlorination by a long-lived intermediate in the mechanism of flavin-dependent halogenases
Biochemistry, 46, 2007
|
|
2OGY
 
 | | Asn199Ala Mutant of the 5-methyltetrahydrofolate corrinoid/iron sulfur protein methyltransferase complexed with methyltetrahydrofolate to 2.3 Angstrom resolution | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-methyltetrahydrofolate corrinoid/iron sulfur protein methyltransferase, CALCIUM ION | | Authors: | Doukov, T.I, Drennan, C.L, Hemmi, H, Ragsdale, S.W. | | Deposit date: | 2007-01-09 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic evidence for an extended hydrogen-bonding network in catalysis of methyl group transfer. Role of an active site asparagine residue in activation of methyl transfer by methyltransferases.
J.Biol.Chem., 282, 2007
|
|
2R0G
 
 | | Chromopyrrolic acid-soaked RebC with bound 7-carboxy-K252c | | Descriptor: | 7-carboxy-5-hydroxy-12,13-dihydro-6H-indolo[2,3-a]pyrrolo[3,4-c]carbazole, FLAVIN-ADENINE DINUCLEOTIDE, RebC | | Authors: | Ryan, K.S, Drennan, C.L. | | Deposit date: | 2007-08-19 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystallographic trapping in the rebeccamycin biosynthetic enzyme RebC
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2QM8
 
 | | MeaB, A Bacterial Homolog of MMAA, in the Nucleotide Free Form | | Descriptor: | GTPase/ATPase, PHOSPHATE ION | | Authors: | Hubbard, P.A, Padovani, D, Labunska, T, Mahlstedt, S.A, Banerjee, R, Drennan, C.L. | | Deposit date: | 2007-07-14 | | Release date: | 2007-08-28 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and mutagenesis of the metallochaperone MeaB: insight into the causes of methylmalonic aciduria.
J.Biol.Chem., 282, 2007
|
|
2E4G
 
 | | RebH with bound L-Trp | | Descriptor: | TRYPTOPHAN, Tryptophan halogenase | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2006-12-07 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Chlorination by a long-lived intermediate in the mechanism of flavin-dependent halogenases
Biochemistry, 46, 2007
|
|
2E7F
 
 | | 5-methyltetrahydrofolate corrinoid/iron sulfur protein methyltransferase complexed with methyltetrahydrofolate to 2.2 Angsrom resolution | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-methyltetrahydrofolate corrinoid/iron sulfur protein methyltransferase, CALCIUM ION | | Authors: | Doukov, T.I, Drennan, C.L, Hemmi, H, Ragsdale, S.W. | | Deposit date: | 2007-01-09 | | Release date: | 2007-01-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and kinetic evidence for an extended hydrogen-bonding network in catalysis of methyl group transfer. Role of an active site asparagine residue in activation of methyl transfer by methyltransferases.
J.Biol.Chem., 282, 2007
|
|
2FCV
 
 | | SyrB2 with Fe(II), bromide, and alpha-ketoglutarate | | Descriptor: | ((2R,3S,4S,5S)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)-5-((2R,3S,4S,5S,6R)-3,4,5-TRIHYDROXY-6-METHOXY-TETRAHYDRO-2H-PYRAN-2-YLOXY)-TETRAHYDROFURAN-2-YL)METHYL NONANOATE, 2-OXOGLUTARIC ACID, BROMIDE ION, ... | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2005-12-12 | | Release date: | 2006-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis.
Nature, 440, 2006
|
|
