7TBU
 
 | | Crystal structure of the 5-enolpyruvate-shikimate-3-phosphate synthase (EPSPS) domain of Aro1 from Candida albicans in complex with shikimate-3-phosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-enolpyruvylshikimate-3-phosphate synthase, SHIKIMATE-3-PHOSPHATE | | Authors: | Stogios, P.J, Evdokimova, E, Michalska, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular analysis and essentiality of Aro1 shikimate biosynthesis multi-enzyme in Candida albicans.
Life Sci Alliance, 5, 2022
|
|
6DM4
 
 | | Crystal structure of the SH2 domain from RavO (Lpg1129) from Legionella pneumophila in complex with Homo sapiens Shc1 phospho-Tyr317 peptide | | Descriptor: | RavO, SULFATE ION, Shc1 phospho-Tyr317 peptide | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Kaneko, T, Li, S, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-06-04 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the SH2 domain from RavO (Lpg1129) from Legionella pneumophila in complex with Homo sapiens Shc1 phospho-Tyr317 peptide
To Be Published
|
|
5VU3
 
 | | Crystal structure of the competence-damaged protein (CinA) superfamily protein ECL_02051 from Enterobacter cloacae | | Descriptor: | ACETATE ION, Competence damage-inducible protein A, TETRAETHYLENE GLYCOL | | Authors: | Stogios, P.J, Skarina, T, McChesney, C, Sandoval, J, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-05-18 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | Crystal structure of the competence-damaged protein (CinA) superfamily protein ECL_02051 from Enterobacter cloacae
To Be Published
|
|
5VM2
 
 | | Crystal structure of ECK1772, an oxidoreductase/dehydrogenase of unknown specificity involved in membrane biogenesis from Escherichia coli | | Descriptor: | Alcohol dehydrogenase, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Stogios, P.J, Skarina, T, McChesney, C, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-26 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Crystal structure of ECK1772, an oxidoreductase/dehydrogenase of unknown specificity involved in membrane biogenesis from Escherichia coli
To Be Published
|
|
6DM3
 
 | | Crystal structure of the SH2 domain from RavO (Lpg1129) from Legionella pneumophila, apoprotein | | Descriptor: | RavO | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Kaneko, T, Li, S, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-06-04 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the SH2 domain from RavO (Lpg1129) from Legionella pneumophila, apoprotein
To Be Published
|
|
7TI9
 
 | | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2, form 2 | | Descriptor: | CHLORIDE ION, GLYCEROL, Papain-like protease nsp3 | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-13 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal structure of the ubiquitin-like domain 1 (Ubl1) of Nsp3 from SARS-CoV-2, form 2
To Be Published
|
|
6D2Y
 
 | | Crystal structure of surface glycan-binding protein PbSGBP-B from Prevotella bryantii | | Descriptor: | GLYCEROL, MAGNESIUM ION, PbSGBP-B lipoprotein | | Authors: | Stogios, P.J, Skarina, T, Wawrzak, Z, McGregor, N, Di Leo, R, Brumer, H, Savchenko, A. | | Deposit date: | 2018-04-14 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of surface glycan-binding protein PbSGBP-B from Prevotella bryantii
To Be Published
|
|
7T9W
 
 | | Crystal structure of the Nsp3 bSM (Betacoronavirus-Specific Marker) domain from SARS-CoV-2 | | Descriptor: | CHLORIDE ION, GLYCEROL, Papain-like protease nsp3 | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-20 | | Release date: | 2021-12-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Nsp3 bSM (Betacoronavirus-Specific Marker) domain from SARS-CoV-2
To Be Published
|
|
7UXG
 
 | | Crystal structure of putative serine protease YdgD from Escherichia coli | | Descriptor: | Serine protease | | Authors: | Stogios, P.J, Michalska, K, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-05-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structure of putative serine protease YdgD from Escherichia coli
To Be Published
|
|
7V09
 
 | | Crystal structure of ECL_RS08780, putative sugar transport system periplasmic sugar-binding protein | | Descriptor: | MAGNESIUM ION, Multiple sugar transport system periplasmic sugar-binding protein | | Authors: | Stogios, P.J, Skarina, T, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-05-10 | | Release date: | 2022-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of ECL_RS08780, putative sugar transport system periplasmic sugar-binding protein
To Be Published
|
|
5UXA
 
 | | Crystal structure of macrolide 2'-phosphotransferase MphB from Escherichia coli | | Descriptor: | CALCIUM ION, Macrolide 2'-phosphotransferase II | | Authors: | Stogios, P.J, Evdokimova, E, Egorova, O, Di Leo, R, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
6HCD
 
 | | Structure of universal stress protein from Archaeoglobus fulgidus | | Descriptor: | ACETATE ION, CHLORIDE ION, UNIVERSAL STRESS PROTEIN, ... | | Authors: | Shumilin, I.A, Loch, J.I, Cymborowski, M, Xu, X, Edwards, A, Di Leo, R, Shabalin, I.G, Joachimiak, A, Savchenko, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-08-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insight into the universal stress protein family.
Evol Appl, 6, 2013
|
|
7JH3
 
 | | Crystal structure of 4-aminobutyrate aminotransferase PuuE from Escherichia coli in complex with PLP | | Descriptor: | 4-aminobutyrate aminotransferase PuuE, DI(HYDROXYETHYL)ETHER | | Authors: | Valleau, D, Evdokimova, E, Stogios, P.J, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-20 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of 4-aminobutyrate aminotransferase PuuE from Escherichia coli in complex with PLP
To Be Published
|
|
4ZMH
 
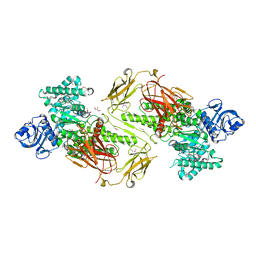 | | Crystal structure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T | | Descriptor: | ACETATE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Nocek, B, Cui, H, Wang, W, Savchenko, A. | | Deposit date: | 2015-05-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Biochemical and Structural Characterization of a Five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T.
J.Biol.Chem., 291, 2016
|
|
6PI9
 
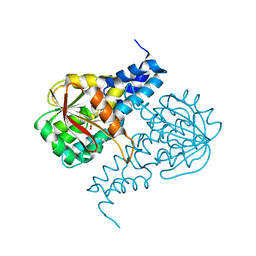 | | Crystal structure of 16S rRNA methyltransferase RmtF in complex with S-Adenosyl-L-homocysteine | | Descriptor: | 16S rRNA (guanine(1405)-N(7))-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Stogios, P.J, Kim, Y, Evdokimova, E, Di Leo, R, Semper, C, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-06-26 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of 16S rRNA methylase RmtF in complex with S-Adenosyl-L-homocysteine
To be Published
|
|
8EZR
 
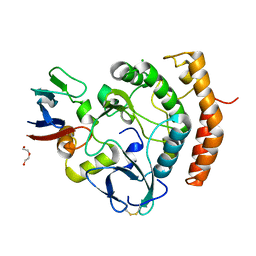 | | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, native protein | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, HipS(Lp), ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Di Leo, R, Lin, J, Ensminger, A, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-11-01 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, native protein
To Be Published
|
|
6PTA
 
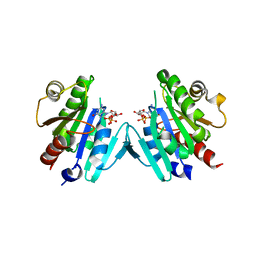 | | Crystal structure of the ARF family small GTPase ARF1 from Candida albicans in complex with GDP | | Descriptor: | ADP-ribosylation factor, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-15 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the ARF family small GTPase ARF1 from Candida albicans in complex with GDP
To Be Published
|
|
8EJU
 
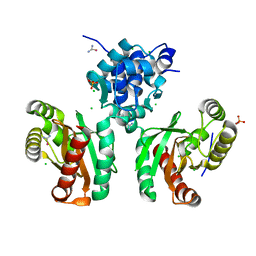 | | The crystal structure of Pseudomonas putida PcaR | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Transcription regulatory protein (Pca regulon), ... | | Authors: | Pham, C, Skarina, T, Di Leo, R, Stogios, P.J, Mahadevan, R, Savchenko, A. | | Deposit date: | 2022-09-19 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The crystal structure of Pseudomonas putida PcaR
To Be Published
|
|
8EJV
 
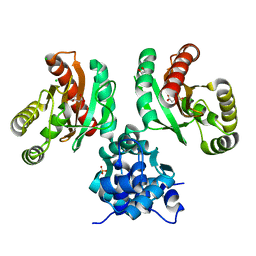 | | The crystal structure of Pseudomonas putida PcaR in complex with succinate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Pham, C, Skarina, T, Di Leo, R, Stogios, P.J, Mahadevan, R, Savchenko, A. | | Deposit date: | 2022-09-19 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The crystal structure of Pseudomonas putida PcaR in complex with succinate
To Be Published
|
|
7RCA
 
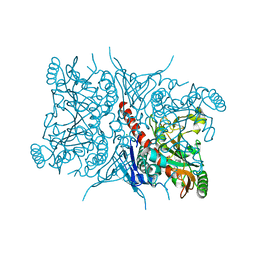 | | Crystal structure of Aro2p chorismate synthase from Candida lusitaniae | | Descriptor: | CHLORIDE ION, Chorismate synthase, SULFATE ION | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-07 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of Aro2p chorismate synthase from Candida lusitaniae
To Be Published
|
|
7RGE
 
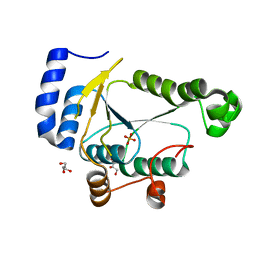 | | Crystal structure of phosphoadenylyl-sulfate (PAPS) reductase from Candida auris, phosphate complex | | Descriptor: | 3'-phosphoadenylylsulfate reductase, GLYCEROL, PHOSPHATE ION | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of phosphoadenylyl-sulfate (PAPS) reductase from Candida auris, phosphate complex
To Be Published
|
|
7RGN
 
 | | Crystal structure of putative fructose-1,6-bisphosphate aldolase from Candida auris | | Descriptor: | Fructose-bisphosphate aldolase, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Tan, K, Di Leo, R, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative fructose-1,6-bisphosphate aldolase from Candida auris
To Be Published
|
|
7RH8
 
 | | Crystal structure of Fur1p from Candida albicans, in complex with UTP | | Descriptor: | URIDINE 5'-TRIPHOSPHATE, Uracil phosphoribosyltransferase | | Authors: | Stogios, P.J, Skarina, T, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-16 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of Fur1p from Candida albicans, in complex with UTP
To Be Published
|
|
7RJ1
 
 | | Crystal structure of Aro7p chorismate mutase from Candida albicans, complex with L-Trp | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Chorismate mutase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Tan, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-20 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of Aro7p chorismate mutase from Candida albicans, complex with L-Trp
To Be Published
|
|
7REU
 
 | | Crystal structure of Aro4p, 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase from Candida auris, L-Tyr complex | | Descriptor: | 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Tan, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-13 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of Aro4p, 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase from Candida auris, L-Tyr complex
To Be Published
|
|
