7QPW
 
 | |
7QFA
 
 | |
7QM2
 
 | |
7QF7
 
 | |
7QFB
 
 | | Crystal structure of Protein Phosphatase 1 in complex with PP1-binding peptide from PTG | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Protein phosphatase 1 regulatory subunit 3C, ... | | Authors: | Semrau, M.S, Storici, P, Lolli, G. | | Deposit date: | 2021-12-05 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular architecture of the glycogen- committed PP1/PTG holoenzyme.
Nat Commun, 13, 2022
|
|
7QQ0
 
 | |
7QPZ
 
 | |
7QQ1
 
 | |
7QPY
 
 | |
6ZLB
 
 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with Indole-3-carbinol | | Descriptor: | 1~{H}-indol-3-ylmethanol, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Fata, F, Silvestri, I, Williams, D.L, Angelucci, F. | | Deposit date: | 2020-06-30 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the Surface of a Parasite Drug Target Thioredoxin Glutathione Reductase Using Small Molecule Fragments.
Acs Infect Dis., 7, 2021
|
|
6ZST
 
 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with 3-(3-methoxyquinoxalin-2-yl)propanoic acid | | Descriptor: | 3-(3-methoxyquinoxalin-2-yl)propanoic acid, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Fata, F, Silvestri, I, Williams, D.L, Angelucci, F. | | Deposit date: | 2020-07-16 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the Surface of a Parasite Drug Target Thioredoxin Glutathione Reductase Using Small Molecule Fragments.
Acs Infect Dis., 7, 2021
|
|
6ZLP
 
 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with 4-Aminopiazthiole | | Descriptor: | 2,1,3-benzothiadiazol-4-amine, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Fata, F, Silvestri, I, Williams, D.L, Angelucci, F. | | Deposit date: | 2020-06-30 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Probing the Surface of a Parasite Drug Target Thioredoxin Glutathione Reductase Using Small Molecule Fragments.
Acs Infect Dis., 7, 2021
|
|
6ZP3
 
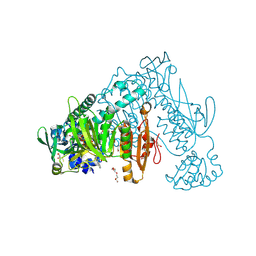 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with 2-Methylindole-3-acetic acid | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(2-methyl-1~{H}-indol-3-yl)ethanoic acid, CALCIUM ION, ... | | Authors: | Fata, F, Silvestri, I, Williams, D.L, Angelucci, F. | | Deposit date: | 2020-07-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the Surface of a Parasite Drug Target Thioredoxin Glutathione Reductase Using Small Molecule Fragments.
Acs Infect Dis., 7, 2021
|
|
7B02
 
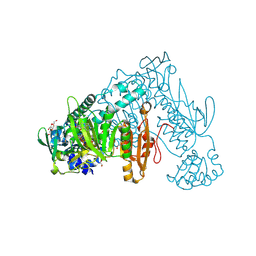 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with 4-Hydroxy-7-methyl-1,8-naphthyridine-3-carboxylic acid | | Descriptor: | 4-Hydroxy-7-methyl-1,8-naphthyridine-3-carboxylic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fata, F, Silvestri, I, Williams, D.L, Angelucci, F. | | Deposit date: | 2020-11-18 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Probing the Surface of a Parasite Drug Target Thioredoxin Glutathione Reductase Using Small Molecule Fragments.
Acs Infect Dis., 7, 2021
|
|
7AHW
 
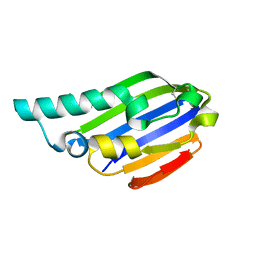 | |
7ALY
 
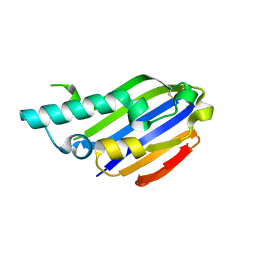 | |
7AMX
 
 | |
8A5W
 
 | | Crystal structure of the human phosphoserine aminotransferase (PSAT) in complex with O-phosphoserine | | Descriptor: | (2S)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-3-phosphonooxy-propanoic acid, (2~{S})-2-[[(~{R})-[[(5~{S})-5-azanyl-6-oxidanylidene-hexyl]amino]-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methyl]amino]-3-phosphonooxy-propanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Costanzi, E, Demitri, N, Ullah, R, Marchesan, F, Peracchi, A, Zangelmi, E, Storici, P, Campanini, B. | | Deposit date: | 2022-06-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | L-serine biosynthesis in the human central nervous system: Structure and function of phosphoserine aminotransferase.
Protein Sci., 32, 2023
|
|
8A5V
 
 | | Crystal structure of the human phosposerine aminotransferase (PSAT) | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Phosphoserine aminotransferase, SULFATE ION | | Authors: | Costanzi, E, Demitri, N, Ullah, R, Marchesan, F, Peracchi, A, Zangelmi, E, Storici, P, Campanini, B. | | Deposit date: | 2022-06-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | L-serine biosynthesis in the human central nervous system: Structure and function of phosphoserine aminotransferase.
Protein Sci., 32, 2023
|
|
8OKL
 
 | | Crystal structure of F2F-2020185-01X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-03-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
8OKM
 
 | | Crystal structure of F2F-2020197-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-03-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
8OKK
 
 | | Crystal structure of F2F-2020184-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-03-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
8OKN
 
 | | Crystal structure of F2F-2020198-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-03-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
7Q5E
 
 | | Crystal structure of F2F-2020209-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, SODIUM ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2021-11-03 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Easy access to alpha-ketoamides as SARS-CoV-2 and MERS M pro inhibitors via the PADAM oxidation route.
Eur.J.Med.Chem., 244, 2022
|
|
7Q5F
 
 | | Crystal structure of F2F-2020216-01X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | (S)-1-(2-(2,4-dichlorophenoxy)acetyl)-N-((S)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)-4-(phenethylamino)butan-2-yl)pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, 3C-like proteinase, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2021-11-03 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Easy access to alpha-ketoamides as SARS-CoV-2 and MERS M pro inhibitors via the PADAM oxidation route.
Eur.J.Med.Chem., 244, 2022
|
|
