7TDU
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-1 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxo(1-~2~H)pyrrolidin-3-yl]propan-2-yl}-3-{N-[tert-butyl(~2~H)carbamoyl]-3-methyl-L-(N-~2~H)valyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-(~2~H)carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
6D54
 
 | |
6E21
 
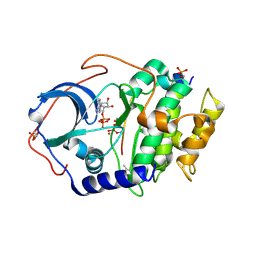 | | Joint X-ray/neutron structure of PKAc with products Sr2-ADP and phosphorylated peptide SP20 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, STRONTIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A, Gerlits, O.O, Taylor, S. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Zooming in on protons: Neutron structure of protein kinase A trapped in a product complex.
Sci Adv, 5, 2019
|
|
5KWF
 
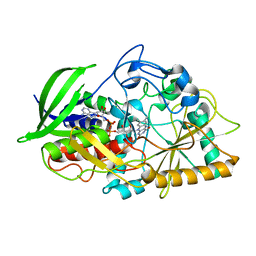 | | Joint X-ray Neutron Structure of Cholesterol Oxidase | | Descriptor: | Cholesterol oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Golden, E, Vrielink, A, Meilleur, F, Blakeley, M. | | Deposit date: | 2016-07-18 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.499 Å), X-RAY DIFFRACTION | | Cite: | An extended N-H bond, driven by a conserved second-order interaction, orients the flavin N5 orbital in cholesterol oxidase.
Sci Rep, 7, 2017
|
|
6KMP
 
 | | 100K X-ray structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with tetrahedral intermediate mimic KVS-1 | | Descriptor: | N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide, Protease | | Authors: | Das, A, Kovalevsky, A. | | Deposit date: | 2019-07-31 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Visualizing Tetrahedral Oxyanion Bound in HIV-1 Protease Using Neutrons: Implications for the Catalytic Mechanism and Drug Design.
Acs Omega, 5, 2020
|
|
8EYS
 
 | |
8EZC
 
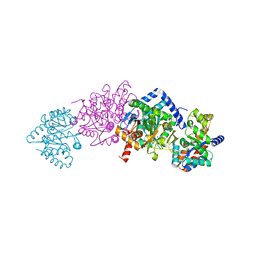 | |
4JEC
 
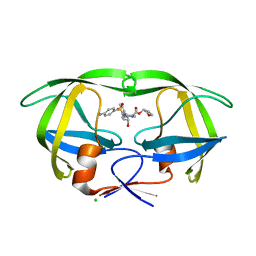 | | Joint neutron and X-ray structure of per-deuterated HIV-1 protease in complex with clinical inhibitor amprenavir | | Descriptor: | CHLORIDE ION, HIV-1 protease, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Kovalevsky, A.Y, Weber, I.T, Langan, P. | | Deposit date: | 2013-02-26 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2.01 Å), X-RAY DIFFRACTION | | Cite: | Joint X-ray/Neutron Crystallographic Study of HIV-1 Protease with Clinical Inhibitor Amprenavir: Insights for Drug Design.
J.Med.Chem., 56, 2013
|
|
3KYU
 
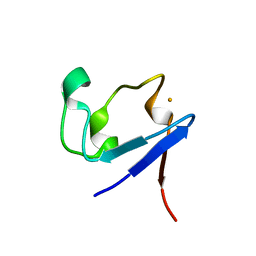 | |
3KYV
 
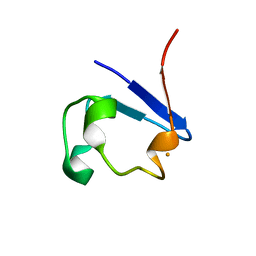 | |
3KYX
 
 | |
3KYW
 
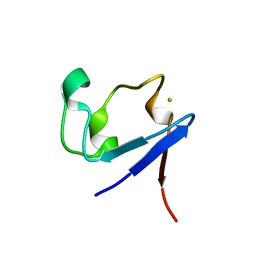 | |
3KYY
 
 | |
7SI9
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with PF-07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-10-12 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TUR
 
 | | Joint X-ray/neutron structure of aspastate aminotransferase (AAT) in complex with pyridoxamine 5'-phosphate (PMP) | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase, ... | | Authors: | Drago, V.N, Kovalevsky, A.Y, Dajnowicz, S, Mueser, T.C. | | Deposit date: | 2022-02-03 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | An N⋯H⋯N low-barrier hydrogen bond preorganizes the catalytic site of aspartate aminotransferase to facilitate the second half-reaction.
Chem Sci, 13, 2022
|
|
5JQR
 
 | | The Structure of Ascorbate Peroxidase Compound II formed by reaction with m-CPBA | | Descriptor: | Ascorbate peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kwon, H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2016-05-05 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Direct visualization of a Fe(IV)-OH intermediate in a heme enzyme.
Nat Commun, 7, 2016
|
|
5E5K
 
 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with darunavir at pH 4.3 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 protease | | Authors: | Kovalevsky, A.Y, Das, A. | | Deposit date: | 2015-10-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.75 Å), X-RAY DIFFRACTION | | Cite: | Long-Range Electrostatics-Induced Two-Proton Transfer Captured by Neutron Crystallography in an Enzyme Catalytic Site.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5E5J
 
 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with darunavir at pH 6.0 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Kovalevsky, A.Y, Gerlits, O.O. | | Deposit date: | 2015-10-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Long-Range Electrostatics-Induced Two-Proton Transfer Captured by Neutron Crystallography in an Enzyme Catalytic Site.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
6PU8
 
 | | Room temperature X-ray structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with tetrahedral intermediate of keto-darunavir | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3,3-dihydroxy-1-phenylbutan-2-yl]carbamate, HIV-1 protease | | Authors: | Kovalevsky, A, Das, A. | | Deposit date: | 2019-07-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Visualizing Tetrahedral Oxyanion Bound in HIV-1 Protease Using Neutrons: Implications for the Catalytic Mechanism and Drug Design.
Acs Omega, 5, 2020
|
|
6PTP
 
 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with tetrahedral intermediate mimic KVS-1 | | Descriptor: | HIV-1 Protease, N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide | | Authors: | Kovalevsky, A, Das, A. | | Deposit date: | 2019-07-16 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Visualizing Tetrahedral Oxyanion Bound in HIV-1 Protease Using Neutrons: Implications for the Catalytic Mechanism and Drug Design.
Acs Omega, 5, 2020
|
|
7NRW
 
 | | Human myelin protein P2 mutant M114T | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Myelin P2 protein, ... | | Authors: | Uusitalo, M, Ruskamo, S, Kursula, P. | | Deposit date: | 2021-03-04 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human myelin protein P2: from crystallography to time-lapse membrane imaging and neuropathy-associated variants.
Febs J., 288, 2021
|
|
7NSR
 
 | | Myelin protein P2 I50del | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Myelin P2 protein, ... | | Authors: | Uusitalo, M, Ruskamo, S, Kursula, P. | | Deposit date: | 2021-03-08 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Human myelin protein P2: from crystallography to time-lapse membrane imaging and neuropathy-associated variants.
Febs J., 288, 2021
|
|
2YCG
 
 | |
7RB5
 
 | | Room temperature structure of hAChE in complex with substrate analog 4K-TMA | | Descriptor: | 4,4-DIHYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, Acetylcholinesterase | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2021-07-05 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Room temperature crystallography of human acetylcholinesterase bound to a substrate analogue 4K-TMA: Towards a neutron structure
Curr Res Struct Biol, 3, 2021
|
|
7RB6
 
 | | Low temperature structure of hAChE in complex with substrate analog 4K-TMA | | Descriptor: | 4,4-DIHYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2021-07-05 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Room temperature crystallography of human acetylcholinesterase bound to a substrate analogue 4K-TMA: Towards a neutron structure
Curr Res Struct Biol, 3, 2021
|
|
