3QOM
 
 | | Crystal structure of 6-phospho-beta-glucosidase from Lactobacillus plantarum | | Descriptor: | 6-phospho-beta-glucosidase, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Michalska, K, Hatzos-Skintges, C, Bearden, J, Kohler, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-10 | | Release date: | 2011-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3QOK
 
 | |
3PMM
 
 | | The crystal structure of a possible member of GH105 family from Klebsiella pneumoniae subsp. pneumoniae MGH 78578 | | Descriptor: | FORMIC ACID, IMIDAZOLE, Putative cytoplasmic protein | | Authors: | Tan, K, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-17 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | The crystal structure of a possible member of GH105 family from Klebsiella pneumoniae subsp. pneumoniae MGH 78578
To be Published
|
|
3PN8
 
 | | The crystal structure of 6-phospho-beta-glucosidase from Streptococcus mutans UA159 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, Putative phospho-beta-glucosidase, ... | | Authors: | Tan, K, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-18 | | Release date: | 2010-12-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | The crystal structure of 6-phospho-beta-glucosidase from Streptococcus mutants UA159
To be Published
|
|
3QSG
 
 | |
3QSJ
 
 | | Crystal structure of NUDIX hydrolase from Alicyclobacillus acidocaldarius | | Descriptor: | CALCIUM ION, GLYCEROL, NUDIX hydrolase | | Authors: | Michalska, K, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-21 | | Release date: | 2011-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of NUDIX hydrolase from Alicyclobacillus acidocaldarius
To be Published
|
|
3O6P
 
 | | Crystal structure of peptide ABC transporter, peptide-binding protein | | Descriptor: | Peptide ABC transporter, peptide-binding protein, SODIUM ION | | Authors: | Chang, C, Bigelow, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-29 | | Release date: | 2010-09-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of peptide ABC transporter, peptide-binding protein
To be Published
|
|
3R89
 
 | |
3RHT
 
 | | Crystal structure of type 1 glutamine amidotransferase (GATase1)-like protein from Planctomyces limnophilus | | Descriptor: | (GATase1)-like protein, ACETATE ION, CALCIUM ION, ... | | Authors: | Michalska, K, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-12 | | Release date: | 2011-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of type 1 glutamine amidotransferase (GATase1)-like protein from Planctomyces limnophilus
To be Published
|
|
3RMS
 
 | | Crystal structure of uncharacterized protein Svir_20580 from Saccharomonospora viridis | | Descriptor: | GLYCEROL, ZINC ION, uncharacterized protein | | Authors: | Michalska, K, Weger, A, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-21 | | Release date: | 2011-05-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Crystal structure of uncharacterized protein Svir_20580 from Saccharomonospora viridis
To be Published
|
|
3RQ0
 
 | | The crystal structure of a glycosyl hydrolases (GH) family protein 16 from Mycobacterium smegmatis str. MC2 155 | | Descriptor: | 2,2',2''-NITRILOTRIETHANOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, K, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-05-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The crystal structure of a glycosyl hydrolases (GH) family protein 16 from Mycobacterium smegmatis str. MC2 155
To be Published
|
|
3RMQ
 
 | | Crystal structure of uncharacterized protein Svir_20580 from Saccharomonospora viridis (V71M mutant) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ZINC ION, ... | | Authors: | Michalska, K, Weger, A, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-21 | | Release date: | 2011-05-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of uncharacterized protein Svir_20580 from Saccharomonospora viridis (V71M mutant)
To be Published
|
|
3QWT
 
 | |
3RJT
 
 | | Crystal structure of lipolytic protein G-D-S-L family from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Chang, C, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-15 | | Release date: | 2011-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of lipolytic protein G-D-S-L family from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446
To be Published
|
|
3RQB
 
 | |
3RNL
 
 | | Crystal Structure of Sulfotransferase from Alicyclobacillus acidocaldarius | | Descriptor: | GLYCEROL, MAGNESIUM ION, Sulfotransferase | | Authors: | Kim, Y, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-22 | | Release date: | 2011-06-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Crystal Structure of Sulfotransferase from Alicyclobacillus acidocaldarius
To be Published
|
|
3ROB
 
 | |
3RXZ
 
 | | Crystal structure of putative polysaccharide deacetylase from Mycobacterium smegmatis | | Descriptor: | CHLORIDE ION, Polysaccharide deacetylase, ZINC ION | | Authors: | Michalska, K, Tesar, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-05-10 | | Release date: | 2011-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of putative polysaccharide deacetylase from Mycobacterium smegmatis
To be Published
|
|
3OGH
 
 | | Crystal structure of yciE protein from E. coli CFT073, a member of ferritine-like superfamily of diiron-containing four-helix-bundle proteins | | Descriptor: | CHLORIDE ION, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Nocek, B, Bigelow, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-16 | | Release date: | 2010-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of yciE protein from E. coli CFT073, a member of ferritine-like superfamily of diiron-containing four-helix-bundle proteins
To be Published
|
|
3OIR
 
 | | Crystal structure of sulfate transporter family protein from Wolinella succinogenes | | Descriptor: | CHLORIDE ION, FORMIC ACID, SULFATE TRANSPORTER SULFATE TRANSPORTER FAMILY PROTEIN | | Authors: | Chang, C, Tesar, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-19 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of sulfate transporter family protein from Wolinella succinogenes
To be Published
|
|
4DGH
 
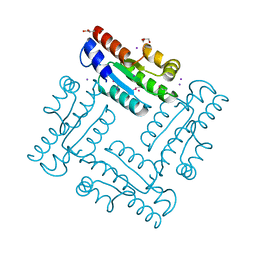 | | Structure of SulP Transporter STAS Domain from Vibrio Cholerae Refined to 1.9 Angstrom Resolution | | Descriptor: | GLYCEROL, IODIDE ION, POTASSIUM ION, ... | | Authors: | Keller, J.P, Chang, C, Marshall, N, Bearden, J, Dallos, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-25 | | Release date: | 2012-02-08 | | Last modified: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of SulP Transporter STAS Domain from Vibrio Cholerae Refined to 1.9 Angstrom Resolution
To be Published
|
|
4GPN
 
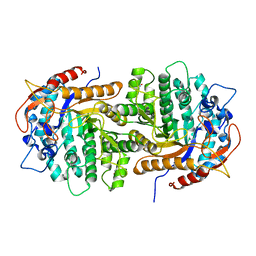 | | The crystal structure of 6-P-beta-D-Glucosidase (E375Q mutant) from Streptococcus mutans UA150 in complex with Gentiobiose 6-phosphate. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-O-phosphono-beta-D-glucopyranose-(1-6)-beta-D-glucopyranose, 6-phospho-beta-D-Glucosidase, ... | | Authors: | Tan, K, Michalska, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
4F79
 
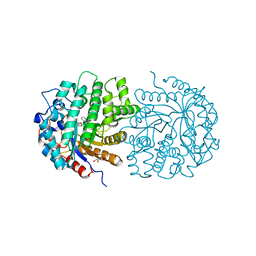 | | The crystal structure of 6-phospho-beta-glucosidase mutant (E375Q) in complex with Salicin 6-phosphate | | Descriptor: | 2-(hydroxymethyl)phenyl 6-O-phosphono-beta-D-glucopyranoside, GLYCEROL, Putative phospho-beta-glucosidase | | Authors: | Tan, K, Michalska, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-15 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
4I4J
 
 | | The structure of SgcE10, the ACP-polyene thioesterase involved in C-1027 biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, ACP-polyene thioesterase, D(-)-TARTARIC ACID, ... | | Authors: | Kim, Y, Bigelow, L, Bearden, J, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-12 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.784 Å) | | Cite: | Crystal Structure of Thioesterase SgcE10 Supporting Common Polyene Intermediates in 9- and 10-Membered Enediyne Core Biosynthesis.
Acs Omega, 2, 2017
|
|
4DGF
 
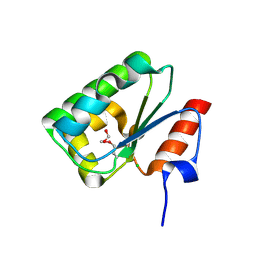 | | Structure of SulP Transporter STAS Domain from Wolinella Succinogenes Refined to 1.6 Angstrom Resolution | | Descriptor: | CHLORIDE ION, FORMIC ACID, SULFATE TRANSPORTER SULFATE TRANSPORTER FAMILY PROTEIN | | Authors: | Keller, J.P, Chang, C, Tesar, C, Bearden, J, Dallos, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-01-25 | | Release date: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of SulP Transporter STAS Domain from Wolinella Succinogenes Refined to 1.6 Angstrom Resolution
To be Published
|
|
