1QQ7
 
 | | STRUCTURE OF L-2-HALOACID DEHALOGENASE FROM XANTHOBACTER AUTOTROPHICUS WITH CHLOROPROPIONIC ACID COVALENTLY BOUND | | Descriptor: | CHLORIDE ION, PROTEIN (L-2-HALOACID DEHALOGENASE) | | Authors: | Ridder, I.S, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1999-06-11 | | Release date: | 1999-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of intermediates in the dehalogenation of haloalkanoates by L-2-haloacid dehalogenase.
J.Biol.Chem., 274, 1999
|
|
1QQ5
 
 | | STRUCTURE OF L-2-HALOACID DEHALOGENASE FROM XANTHOBACTER AUTOTROPHICUS | | Descriptor: | FORMIC ACID, PROTEIN (L-2-HALOACID DEHALOGENASE) | | Authors: | Ridder, I.S, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1999-06-10 | | Release date: | 1999-10-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structures of intermediates in the dehalogenation of haloalkanoates by L-2-haloacid dehalogenase.
J.Biol.Chem., 274, 1999
|
|
7AM3
 
 | | Crystal structure of Peptiligase mutant - M222P | | Descriptor: | GLYCEROL, SULFATE ION, Subtilisin BPN' | | Authors: | Rozeboom, H.J, Janssen, D.J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | From thiol-subtilisin to omniligase: Design and structure of a broadly applicable peptide ligase.
Comput Struct Biotechnol J, 19, 2021
|
|
7AM6
 
 | |
7AM8
 
 | | Crystal structure of Omniligase mutant W189F | | Descriptor: | ACRYLIC ACID, CHLORIDE ION, HISTIDINE, ... | | Authors: | Rozeboom, H.J, Janssen, D.J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | From thiol-subtilisin to omniligase: Design and structure of a broadly applicable peptide ligase.
Comput Struct Biotechnol J, 19, 2021
|
|
7AM4
 
 | |
7AM5
 
 | |
7AM7
 
 | |
2EDC
 
 | |
2EDA
 
 | |
1CIJ
 
 | | HALOALKANE DEHALOGENASE SOAKED WITH HIGH CONCENTRATION OF BROMIDE | | Descriptor: | BROMIDE ION, PROTEIN (HALOALKANE DEHALOGENASE) | | Authors: | Ridder, I.S, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1999-03-31 | | Release date: | 1999-09-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic and kinetic evidence of a collision complex formed during halide import in haloalkane dehalogenase.
Biochemistry, 38, 1999
|
|
1EDB
 
 | |
1EDD
 
 | |
4ZI5
 
 | | Crystal Structure of Dienelactone Hydrolase-like Promiscuous Phospotriesterase P91 from Metagenomic Libraries | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, P91 | | Authors: | Colin, P.-Y, Fischer, G, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2015-04-27 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Ultrahigh-throughput discovery of promiscuous enzymes by picodroplet functional metagenomics.
Nat Commun, 6, 2015
|
|
1QQ6
 
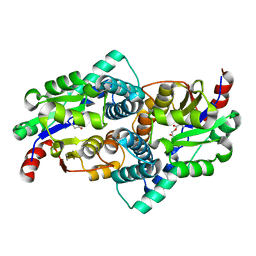 | | STRUCTURE OF L-2-HALOACID DEHALOGENASE FROM XANTHOBACTER AUTOTROPHICUS WITH CHLOROACETIC ACID COVALENTLY BOUND | | Descriptor: | CHLORIDE ION, PROTEIN (L-2-HALOACID DEHALOGENASE) | | Authors: | Ridder, I.S, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1999-06-11 | | Release date: | 1999-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of intermediates in the dehalogenation of haloalkanoates by L-2-haloacid dehalogenase.
J.Biol.Chem., 274, 1999
|
|
8ABS
 
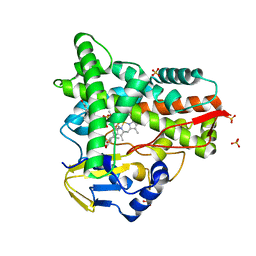 | | Crystal structure of CYP109A2 from Bacillus megaterium bound with testosterone and putative ligand 4,6-dimethyloctanoic acid | | Descriptor: | (4~{S},6~{S})-4,6-dimethyloctanoic acid, Cytochrome P450, HEME B/C, ... | | Authors: | Jozwik, I.K, Rozeboom, H.J, Thunnissen, A.-M.W.H. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Regio- and stereoselective steroid hydroxylation by CYP109A2 from Bacillus megaterium explored by X-ray crystallography and computational modeling.
Febs J., 290, 2023
|
|
8ABR
 
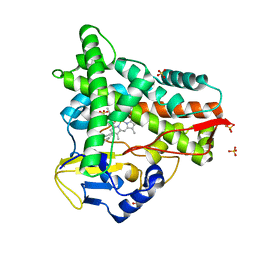 | | Crystal structure of CYP109A2 from Bacillus megaterium bound with putative ligands hexanoic acid and octanoic acid | | Descriptor: | Cytochrome P450, HEME B/C, HEXANOIC ACID, ... | | Authors: | Jozwik, I.K, Rozeboom, H.J, Thunnissen, A.-M.W.H. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Regio- and stereoselective steroid hydroxylation by CYP109A2 from Bacillus megaterium explored by X-ray crystallography and computational modeling.
Febs J., 290, 2023
|
|
2B9V
 
 | | Acetobacter turbidans alpha-amino acid ester hydrolase | | Descriptor: | Alpha-amino acid ester hydrolase | | Authors: | Barends, T.R.M. | | Deposit date: | 2005-10-13 | | Release date: | 2005-12-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Acetobacter turbidans alpha-amino acid ester hydrolase: how a single mutation improves an antibiotic-producing enzyme.
J.Biol.Chem., 281, 2006
|
|
1AQ6
 
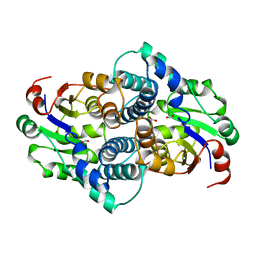 | | STRUCTURE OF L-2-HALOACID DEHALOGENASE FROM XANTHOBACTER AUTOTROPHICUS | | Descriptor: | FORMIC ACID, L-2-HALOACID DEHALOGENASE | | Authors: | Ridder, I.S, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1997-08-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-dimensional structure of L-2-haloacid dehalogenase from Xanthobacter autotrophicus GJ10 complexed with the substrate-analogue formate.
J.Biol.Chem., 272, 1997
|
|
1BEZ
 
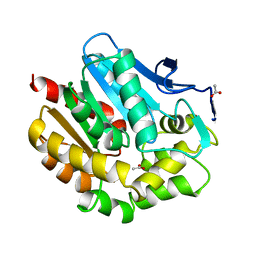 | | HALOALKANE DEHALOGENASE MUTANT WITH TRP 175 REPLACED BY TYR AT PH 5 | | Descriptor: | ACETIC ACID, HALOALKANE DEHALOGENASE | | Authors: | Ridder, I.S, Vos, G.J, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1998-05-18 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic analysis and X-ray structure of haloalkane dehalogenase with a modified halide-binding site.
Biochemistry, 37, 1998
|
|
1BE0
 
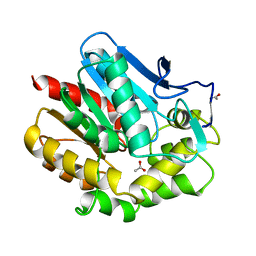 | | HALOALKANE DEHALOGENASE AT PH 5.0 CONTAINING ACETIC ACID | | Descriptor: | ACETATE ION, ACETIC ACID, HALOALKANE DEHALOGENASE | | Authors: | Ridder, I.S, Vos, G.J, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1998-05-18 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Kinetic analysis and X-ray structure of haloalkane dehalogenase with a modified halide-binding site.
Biochemistry, 37, 1998
|
|
1KEC
 
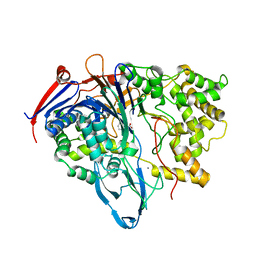 | | PENICILLIN ACYLASE MUTANT WITH PHENYL PROPRIONIC ACID | | Descriptor: | CALCIUM ION, PENICILLIN ACYLASE ALPHA SUBUNIT, PENICILLIN ACYLASE BETA SUBUNIT, ... | | Authors: | Hensgens, C.M.H, Keizer, E, Snijder, H.J, Dijkstra, B.W. | | Deposit date: | 2001-11-15 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic studies on ligand binding in wild-type and active-site mutants of penicillin acylase.
Protein Eng.Des.Sel., 17, 2004
|
|
1BEE
 
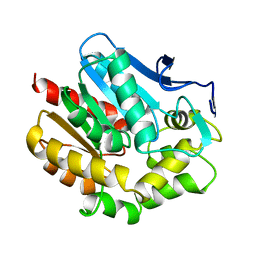 | | HALOALKANE DEHALOGENASE MUTANT WITH TRP 175 REPLACED BY TYR | | Descriptor: | HALOALKANE DEHALOGENASE | | Authors: | Ridder, I.S, Vos, G.J, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1998-05-13 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Kinetic analysis and X-ray structure of haloalkane dehalogenase with a modified halide-binding site.
Biochemistry, 37, 1998
|
|
1K5Q
 
 | | PENICILLIN ACYLASE, MUTANT COMPLEXED WITH PAA | | Descriptor: | 2-PHENYLACETIC ACID, CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, ... | | Authors: | Hensgens, C.M.H, Keizer, E, Snijder, H.J, Dijkstra, B.W. | | Deposit date: | 2001-10-12 | | Release date: | 2003-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural and kinetic studies on ligand binding in wild-type and active-site mutants of penicillin acylase.
Protein Eng.Des.Sel., 17, 2004
|
|
1K5S
 
 | | PENICILLIN ACYLASE, MUTANT COMPLEXED WITH PPA | | Descriptor: | CALCIUM ION, PENICILLIN G ACYLASE ALPHA SUBUNIT, PENICILLIN G ACYLASE BETA SUBUNIT, ... | | Authors: | Hensgens, C.M.H, Keizer, E, Snijder, H.J, Dijkstra, B.W. | | Deposit date: | 2001-10-12 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural and kinetic studies on ligand binding in wild-type and active-site mutants of penicillin acylase.
Protein Eng.Des.Sel., 17, 2004
|
|
