4QKL
 
 | |
1LT1
 
 | |
1COS
 
 | | CRYSTAL STRUCTURE OF A SYNTHETIC TRIPLE-STRANDED ALPHA-HELICAL BUNDLE | | Descriptor: | COILED SERINE | | Authors: | Lovejoy, B, Choe, S, Cascio, D, Mcrorie, D.K, Degrado, W, Eisenberg, D. | | Deposit date: | 1993-01-22 | | Release date: | 1993-10-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a synthetic triple-stranded alpha-helical bundle.
Science, 259, 1993
|
|
1EC5
 
 | | CRYSTAL STRUCTURE OF FOUR-HELIX BUNDLE MODEL | | Descriptor: | PROTEIN (FOUR-HELIX BUNDLE MODEL), ZINC ION | | Authors: | Geremia, S. | | Deposit date: | 2000-01-25 | | Release date: | 2000-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inaugural article: retrostructural analysis of metalloproteins: application to the design of a minimal model for diiron proteins.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1BYZ
 
 | | DESIGNED PEPTIDE ALPHA-1, P1 FORM | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Prive, G.G, Anderson, D.H, Wesson, L, Cascio, D, Eisenberg, D. | | Deposit date: | 1998-10-20 | | Release date: | 1998-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Packed protein bilayers in the 0.90 A resolution structure of a designed alpha helical bundle.
Protein Sci., 8, 1999
|
|
1OVV
 
 | |
1OVU
 
 | |
1OVR
 
 | | CRYSTAL STRUCTURE OF FOUR-HELIX BUNDLE MODEL di-Mn(II)-DF1-L13 | | Descriptor: | MANGANESE (II) ION, four-helix bundle model di-Mn(II)-DF1-L13 | | Authors: | Di Costanzo, L, Geremia, S. | | Deposit date: | 2003-03-27 | | Release date: | 2004-05-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Response of a designed metalloprotein to changes in metal ion coordination, exogenous ligands, and active site volume determined by X-ray crystallography.
J.Am.Chem.Soc., 127, 2005
|
|
2KIK
 
 | |
1JM0
 
 | | CRYSTAL STRUCTURE OF FOUR-HELIX BUNDLE MODEL | | Descriptor: | DIMETHYL SULFOXIDE, MANGANESE (II) ION, PROTEIN (FOUR-HELIX BUNDLE MODEL) | | Authors: | Di Costanzo, L, Geremia, S. | | Deposit date: | 2001-07-17 | | Release date: | 2002-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Toward the de novo design of a catalytically active helix bundle: a substrate-accessible carboxylate-bridged dinuclear metal center.
J.Am.Chem.Soc., 123, 2001
|
|
1JMB
 
 | | CRYSTAL STRUCTURE OF FOUR-HELIX BUNDLE MODEL | | Descriptor: | DIMETHYL SULFOXIDE, MANGANESE (II) ION, PROTEIN (FOUR-HELIX BUNDLE MODEL) | | Authors: | Di Costanzo, L, Geremia, S. | | Deposit date: | 2001-07-18 | | Release date: | 2002-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Toward the de novo design of a catalytically active helix bundle: a substrate-accessible carboxylate-bridged dinuclear metal center.
J.Am.Chem.Soc., 123, 2001
|
|
8FUG
 
 | |
6W6X
 
 | | Crystal Structure of ABLE Apo-protein | | Descriptor: | ACETATE ION, De novo designed ABLE protein, SULFATE ION | | Authors: | Polizzi, N.F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.297 Å) | | Cite: | A defined structural unit enables de novo design of small-molecule-binding proteins.
Science, 369, 2020
|
|
6W70
 
 | | Crystal Structure of apixaban-bound ABLE | | Descriptor: | 1-(4-METHOXYPHENYL)-7-OXO-6-[4-(2-OXOPIPERIDIN-1-YL)PHENYL]-4,5,6,7-TETRAHYDRO-1H-PYRAZOLO[3,4-C]PYRIDINE-3-CARBOXAMIDE, ACETATE ION, De novo designed ABLE, ... | | Authors: | Polizzi, N.F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.296 Å) | | Cite: | A defined structural unit enables de novo design of small-molecule-binding proteins.
Science, 369, 2020
|
|
6X8N
 
 | | Crystal Structure of H49A ABLE mutant | | Descriptor: | De novo designed ABLE protein | | Authors: | Polizzi, N.F. | | Deposit date: | 2020-06-01 | | Release date: | 2020-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A defined structural unit enables de novo design of small-molecule-binding proteins.
Science, 369, 2020
|
|
3URM
 
 | | Crystal structure of the periplasmic sugar binding protein ChvE | | Descriptor: | Multiple sugar-binding periplasmic receptor ChvE, beta-D-galactopyranose | | Authors: | Hu, X, Zhao, J, Binns, A, Degrado, W. | | Deposit date: | 2011-11-22 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3UUG
 
 | | Crystal structure of the periplasmic sugar binding protein ChvE | | Descriptor: | Multiple sugar-binding periplasmic receptor ChvE, beta-D-glucopyranuronic acid | | Authors: | Hu, X, Zhao, J, Binns, A, Degrado, W. | | Deposit date: | 2011-11-28 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3V86
 
 | |
4DAC
 
 | |
2KZ2
 
 | | Calmodulin, C-terminal domain, F92E mutant | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Korendovych, I, Kulp, D, Wu, Y, Cheng, H, Roder, H, DeGrado, W. | | Deposit date: | 2010-06-10 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Design of a switchable eliminase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2LY0
 
 | | Solution NMR structure of the influenza A virus S31N mutant (19-49) in presence of drug M2WJ332 | | Descriptor: | (3S,5S,7S)-N-{[5-(thiophen-2-yl)-1,2-oxazol-3-yl]methyl}tricyclo[3.3.1.1~3,7~]decan-1-aminium, Membrane ion channel M2 | | Authors: | Wu, Y, Wang, J, DeGrado, W. | | Deposit date: | 2012-09-10 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and inhibition of the drug-resistant S31N mutant of the M2 ion channel of influenza A virus.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2MUV
 
 | | NOE-based model of the influenza A virus M2 (19-49) bound to drug 11 | | Descriptor: | (3s,5s,7s)-N-[(5-bromothiophen-2-yl)methyl]tricyclo[3.3.1.1~3,7~]decan-1-aminium, Matrix protein 2 | | Authors: | Wu, Y, Wang, J, DeGrado, W. | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Flipping in the Pore: Discovery of Dual Inhibitors That Bind in Different Orientations to the Wild-Type versus the Amantadine-Resistant S31N Mutant of the Influenza A Virus M2 Proton Channel.
J.Am.Chem.Soc., 136, 2014
|
|
2MUW
 
 | | NOE-based model of the influenza A virus N31S mutant (19-49) bound to drug 11 | | Descriptor: | (3s,5s,7s)-N-[(5-bromothiophen-2-yl)methyl]tricyclo[3.3.1.1~3,7~]decan-1-aminium, Matrix protein 2 | | Authors: | Wu, Y, Wang, J, DeGrado, W. | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Flipping in the Pore: Discovery of Dual Inhibitors That Bind in Different Orientations to the Wild-Type versus the Amantadine-Resistant S31N Mutant of the Influenza A Virus M2 Proton Channel.
J.Am.Chem.Soc., 136, 2014
|
|
5HKN
 
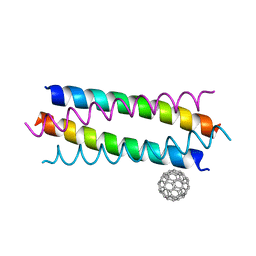 | | Crystal structure de novo designed fullerene organizing protein complex with fullerene | | Descriptor: | (C_{60}-I_{h})[5,6]fullerene, fullerene organizing protein | | Authors: | Paul, J, Acharya, R, Kim, K.-H, Kim, Y.H, Kim, N.H, Grigoryan, G, DeGardo, W.F. | | Deposit date: | 2016-01-14 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Protein-directed self-assembly of a fullerene crystal.
Nat Commun, 7, 2016
|
|
5HKR
 
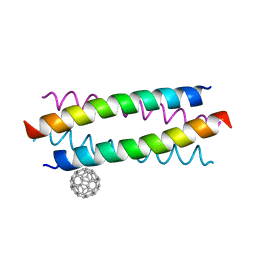 | | Crystal structure of de novo designed fullerene organising protein complex with fullerene | | Descriptor: | (C_{60}-I_{h})[5,6]fullerene, fullerene organizing protein | | Authors: | Acharya, R, Kim, Y.H, Grigoryan, G, DeGardo, W.F. | | Deposit date: | 2016-01-14 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Protein-directed self-assembly of a fullerene crystal.
Nat Commun, 7, 2016
|
|
