3ST2
 
 | | Dreiklang - equilibrium state | | Descriptor: | Dreiklang, PHOSPHATE ION | | Authors: | Brakemann, T, Weber, G, Andresen, M, Stiel, A.C, Jakobs, S, Wahl, M.C. | | Deposit date: | 2011-07-08 | | Release date: | 2011-09-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A reversibly photoswitchable GFP-like protein with fluorescence excitation decoupled from switching.
Nat.Biotechnol., 29, 2011
|
|
5V3Z
 
 | |
7XM5
 
 | | Keap1 Kelch domain (residues 322-609) in complex with 6i | | Descriptor: | Kelch-like ECH-associated protein 1, N-[4-[(2-azanyl-2-oxidanylidene-ethyl)-[4-[(2-azanyl-2-oxidanylidene-ethyl)-(4-methoxyphenyl)sulfonyl-amino]naphthalen-1-yl]sulfamoyl]phenyl]-3-morpholin-4-yl-propanamide | | Authors: | Xu, K. | | Deposit date: | 2022-04-24 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallography-Guided Optimizations of the Keap1-Nrf2 Inhibitors on the Solvent Exposed Region: From Symmetric to Asymmetric Naphthalenesulfonamides.
J.Med.Chem., 65, 2022
|
|
7XM4
 
 | | Crystal structure of Keap1 Kelch domain (residues 322-609) in complex with 6e | | Descriptor: | Kelch-like ECH-associated protein 1, N-[4-[(2-azanyl-2-oxidanylidene-ethyl)-[4-[(2-azanyl-2-oxidanylidene-ethyl)-(4-methoxyphenyl)sulfonyl-amino]naphthalen-1-yl]sulfamoyl]phenyl]-2-(4-ethylpiperazin-1-yl)ethanamide | | Authors: | Xu, K. | | Deposit date: | 2022-04-24 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallography-Guided Optimizations of the Keap1-Nrf2 Inhibitors on the Solvent Exposed Region: From Symmetric to Asymmetric Naphthalenesulfonamides.
J.Med.Chem., 65, 2022
|
|
7XM3
 
 | | Crystal structure of Keap1 Kelch domain (residues 322-609) in complex with 6k | | Descriptor: | Kelch-like ECH-associated protein 1, N-[4-[(2-azanyl-2-oxidanylidene-ethyl)-[4-[(2-azanyl-2-oxidanylidene-ethyl)-(4-methoxyphenyl)sulfonyl-amino]naphthalen-1-yl]sulfamoyl]phenyl]-3-(4-ethylpiperazin-1-yl)propanamide | | Authors: | Xu, K. | | Deposit date: | 2022-04-24 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallography-Guided Optimizations of the Keap1-Nrf2 Inhibitors on the Solvent Exposed Region: From Symmetric to Asymmetric Naphthalenesulfonamides.
J.Med.Chem., 65, 2022
|
|
7XM2
 
 | | Crystal structure of Keap1 Kelch domain (residues 322-609) in complex with NXPZ-2 | | Descriptor: | 2-[(4-aminophenyl)sulfonyl-[4-[(2-azanyl-2-oxidanylidene-ethyl)-(4-methoxyphenyl)sulfonyl-amino]naphthalen-1-yl]amino]ethanamide, Kelch-like ECH-associated protein 1 | | Authors: | Xu, K. | | Deposit date: | 2022-04-24 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallography-Guided Optimizations of the Keap1-Nrf2 Inhibitors on the Solvent Exposed Region: From Symmetric to Asymmetric Naphthalenesulfonamides.
J.Med.Chem., 65, 2022
|
|
5UOS
 
 | | Crystal Structure of CblC (MMACHC) (1-238), a human B12 processing enzyme, complexed with an Antivitamin B12 | | Descriptor: | 1-ethynyl-2,4-difluorobenzene, 2-PHENYLAMINO-ETHANESULFONIC ACID, COBALAMIN, ... | | Authors: | Shanmuganathan, A, Karasik, A, Ruetz, M, Banerjee, R, Krautler, B, Koutmos, M. | | Deposit date: | 2017-02-01 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Antivitamin B12 Inhibition of the Human B12 -Processing Enzyme CblC: Crystal Structure of an Inactive Ternary Complex with Glutathione as the Cosubstrate.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5V42
 
 | |
5V3Y
 
 | |
3ST4
 
 | | Dreiklang - on state | | Descriptor: | Dreiklang, PHOSPHATE ION | | Authors: | Brakemann, T, Weber, G, Andresen, M, Stiel, A.C, Jakobs, S, Wahl, M.C. | | Deposit date: | 2011-07-08 | | Release date: | 2011-09-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A reversibly photoswitchable GFP-like protein with fluorescence excitation decoupled from switching.
Nat.Biotechnol., 29, 2011
|
|
4M08
 
 | | Crystal Structure of Mutant Chlorite Dismutase from Candidatus Nitrospira defluvii W145V | | Descriptor: | 1,2-ETHANEDIOL, Chlorite dismutase, IMIDAZOLE, ... | | Authors: | Gysel, K, Hagmueller, A, Djinovic-Carugo, K. | | Deposit date: | 2013-08-01 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Manipulating conserved heme cavity residues of chlorite dismutase: effect on structure, redox chemistry, and reactivity.
Biochemistry, 53, 2014
|
|
3BO8
 
 | | The High Resolution Crystal Structure of HLA-A1 Complexed with the MAGE-A1 Peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Kumar, P, Vahedi-Faridi, A, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2007-12-17 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational changes within the HLA-A1:MAGE-A1 complex induced by binding of a recombinant antibody fragment with TCR-like specificity
Protein Sci., 18, 2009
|
|
4M07
 
 | | Crystal Structure of Mutant Chlorite Dismutase from Candidatus Nitrospira defluvii W145F | | Descriptor: | 1,2-ETHANEDIOL, Chlorite dismutase, GLYCEROL, ... | | Authors: | Hagmueller, A, Gysel, K, Djinovic-Carugo, K. | | Deposit date: | 2013-08-01 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Manipulating conserved heme cavity residues of chlorite dismutase: effect on structure, redox chemistry, and reactivity.
Biochemistry, 53, 2014
|
|
5V3W
 
 | |
7XMC
 
 | | Cryo-EM structure of Cytochrome bo3 from Escherichia coli, apo structure with DMSO | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Nishida, Y, Shigematsu, H, Iwamoto, T, Takashima, S, Shintani, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
7XMD
 
 | | Cryo-EM structure of Cytochrome bo3 from Escherichia coli, the structure complexed with an allosteric inhibitor N4 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Nishida, Y, Shigematsu, H, Iwamoto, T, Takashima, S, Shintani, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
7XMA
 
 | | Crystal structure of Bovine heart cytochrome c oxidase, apo structure with DMSO | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Nishida, Y, Shinzawa-Itoh, K, Mizuno, N, Kumasaka, T, Yoshikawa, S, Tsukihara, T, Takashima, S, Shintani, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
7XMB
 
 | | Crystal structure of Bovine heart cytochrome c oxidase, the structure complexed with an allosteric inhibitor T113 | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Nishida, Y, Shinzawa-Itoh, K, Mizuno, N, Kumasaka, T, Yoshikawa, S, Tsukihara, T, Shintani, Y, Takashima, S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
1IBR
 
 | | COMPLEX OF RAN WITH IMPORTIN BETA | | Descriptor: | GTP-binding nuclear protein RAN, Importin beta-1 subunit, MAGNESIUM ION, ... | | Authors: | Vetter, I.R, Arndt, A, Kutay, U, Goerlich, D, Wittinghofer, A. | | Deposit date: | 1999-05-14 | | Release date: | 1999-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural view of the Ran-Importin beta interaction at 2.3 A resolution
Cell(Cambridge,Mass.), 97, 1999
|
|
6KDC
 
 | |
5V41
 
 | |
5V3X
 
 | |
2LEV
 
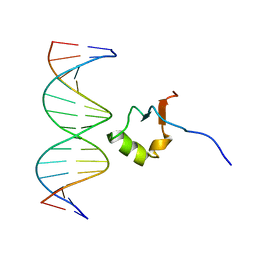 | | Structure of the DNA complex of the C-Terminal domain of Ler | | Descriptor: | DNA (5'-D(*CP*CP*TP*AP*TP*CP*AP*AP*TP*TP*AP*TP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*AP*TP*AP*AP*TP*TP*GP*AP*TP*AP*GP*G)-3'), Ler | | Authors: | Cordeiro, T.N, Schimdt, H, Madrid, C, Juarez, A, Bernado, P, Grisienger, C, Garcia, J, Pons, M. | | Deposit date: | 2011-06-23 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Indirect DNA Readout by an H-NS Related Protein: Structure of the DNA Complex of the C-Terminal Domain of Ler.
Plos Pathog., 7, 2011
|
|
5V40
 
 | |
1VGU
 
 | |
