5UGH
 
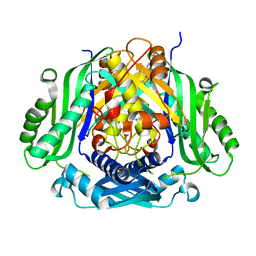 | | Crystal structure of Mat2a bound to the allosteric inhibitor PF-02929366 | | Descriptor: | 2-(7-chloro-5-phenyl[1,2,4]triazolo[4,3-a]quinolin-1-yl)-N,N-dimethylethan-1-amine, S-adenosylmethionine synthase isoform type-2 | | Authors: | Kaiser, S.E, Feng, J, Stewart, A.E. | | Deposit date: | 2017-01-08 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.062 Å) | | Cite: | Targeting S-adenosylmethionine biosynthesis with a novel allosteric inhibitor of Mat2A.
Nat. Chem. Biol., 13, 2017
|
|
4CI9
 
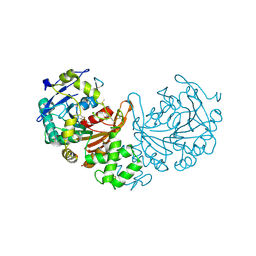 | | Crystal structure of cathepsin A, apo-structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schreuder, H.A, Liesum, A, Kroll, K, Boehnisch, B, Buning, C, Ruf, S, Buning, C, Sadowski, T. | | Deposit date: | 2013-12-06 | | Release date: | 2014-02-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of cathepsin A, a novel target for the treatment of cardiovascular diseases.
Biochem. Biophys. Res. Commun., 445, 2014
|
|
4CIB
 
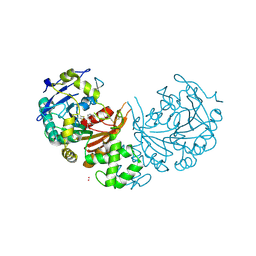 | | crystal structure of cathepsin a, complexed with compound 2 | | Descriptor: | 2-(cyclohexylmethyl)propanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Schreuder, H.A, Liesum, A, Kroll, K, Boehnisch, B, Buning, C, Ruf, S, Buning, C, Sadowski, T. | | Deposit date: | 2013-12-06 | | Release date: | 2014-02-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of cathepsin A, a novel target for the treatment of cardiovascular diseases.
Biochem. Biophys. Res. Commun., 445, 2014
|
|
4CIA
 
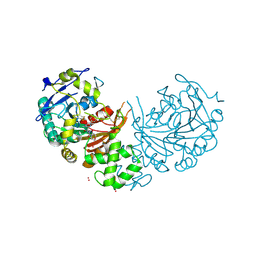 | | Crystal structure of cathepsin A, complexed with compound 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, LYSOSOMAL PROTECTIVE PROTEIN, ... | | Authors: | Schreuder, H.A, Liesum, A, Kroll, K, Boehnisch, B, Buning, C, Ruf, S, Buning, C, Sadowski, T. | | Deposit date: | 2013-12-06 | | Release date: | 2014-02-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of cathepsin A, a novel target for the treatment of cardiovascular diseases.
Biochem. Biophys. Res. Commun., 445, 2014
|
|
2JE6
 
 | | Structure of a 9-subunit archaeal exosome | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, EXOSOME COMPLEX EXONUCLEASE 1, ... | | Authors: | Lorentzen, E, Conti, E. | | Deposit date: | 2007-01-15 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RNA Channelling by the Archaeal Exosome.
Embo Rep., 8, 2007
|
|
2LWB
 
 | | Structural model of BAD-1 repeat loop by NMR | | Descriptor: | Adhesin WI-1 | | Authors: | Brandhorst, T, Klein, B, Tonelli, M. | | Deposit date: | 2012-07-26 | | Release date: | 2013-07-31 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Structure and function of a fungal adhesin that mimics thrombospondin-1 by binding heparin sulfate glycosaminoglycan and suppressing T cell activation via interaction with CD47
To be Published
|
|
2JEA
 
 | |
2JEB
 
 | |
4C46
 
 | | ANDREI-N-LVPAS fused to GCN4 adaptors | | Descriptor: | BROMIDE ION, GENERAL CONTROL PROTEIN GCN4 | | Authors: | Albrecht, R, Alva, V, Ammelburg, M, Baer, K, Basina, E, Boichenko, I, Bonhoeffer, F, Braun, V, Chaubey, M, Chauhan, N, Chellamuthu, V.R, Coles, M, Deiss, S, Ewers, C.P, Forouzan, D, Fuchs, A, Groemping, Y, Hartmann, M.D, Hernandez Alvarez, B, Jeganantham, A, Kalev, I, Koenninger, U, Koiwai, K, Kopec, K.O, Korycinski, M, Laudenbach, B, Lehmann, K, Leo, J.C, Linke, D, Marialke, J, Martin, J, Mechelke, M, Michalik, M, Noll, A, Patzer, S.I, Scharfenberg, F, Schueckel, M, Shahid, S.A, Sulz, E, Ursinus, A, Wuertenberger, S, Zhu, H. | | Deposit date: | 2013-08-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Your Personalized Protein Structure: Andrei N. Lupas Fused to GCN4 Adaptors.
J.Struct.Biol., 186, 2014
|
|
2NOJ
 
 | | Crystal structure of Ehp / C3d complex | | Descriptor: | Complement C3, Efb homologous protein | | Authors: | Hammel, M, Geisbrecht, B.V. | | Deposit date: | 2006-10-25 | | Release date: | 2007-08-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterization of Ehp, a Secreted Complement Inhibitory Protein from Staphylococcus aureus.
J.Biol.Chem., 282, 2007
|
|
