4HDE
 
 | | The crystal structure of a SCO1/SenC family lipoprotein from Bacillus anthracis str. Ames | | Descriptor: | SCO1/SenC family lipoprotein | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-10-02 | | Release date: | 2012-10-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.317 Å) | | Cite: | The crystal structure of a SCO1/SenC family lipoprotein from Bacillus anthracis str. Ames
To be Published
|
|
4H59
 
 | | Crystal structure of iron uptake ABC transporter substrate-binding protein PiaA from Streptococcus pneumoniae Canada MDR_19A bound to Bis-tris propane | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Minasov, G, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-18 | | Release date: | 2012-10-03 | | Last modified: | 2016-05-04 | | Method: | X-RAY DIFFRACTION (1.658 Å) | | Cite: | Crystal structure of iron uptake ABC transporter substrate-binding protein PiaA from Streptococcus pneumoniae Canada MDR_19A bound to Bis-tris propane
To be Published
|
|
4GUH
 
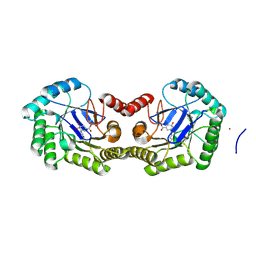 | | 1.95 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) E86A Mutant in Complex with Dehydroshikimate (Crystal Form #2) | | Descriptor: | (4S,5R)-4,5-dihydroxy-3-oxocyclohex-1-ene-1-carboxylic acid, 3-dehydroquinate dehydratase, NICKEL (II) ION | | Authors: | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Reassessing the type I dehydroquinate dehydratase catalytic triad: Kinetic and structural studies of Glu86 mutants.
Protein Sci., 22, 2013
|
|
4HCF
 
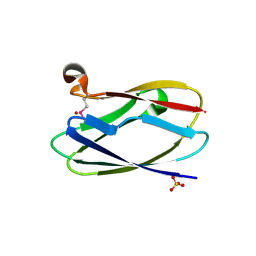 | | Crystal Structure of Uncharacterized Cupredoxin-like Domain Protein Cupredoxin_1 with Copper Bound from Bacillus anthracis | | Descriptor: | COPPER (II) ION, Cupredoxin 1, SULFATE ION | | Authors: | Kim, Y, Maltseva, N, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-29 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Crystal Structure of Uncharacterized Cupredoxin-like Domain Protein Cupredoxin_1 with Copper Bound from Bacillus anthracis
To be Published
|
|
3R7T
 
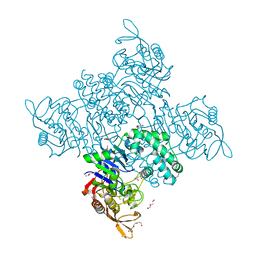 | | Crystal Structure of Adenylosuccinate Synthetase from Campylobacter jejuni | | Descriptor: | Adenylosuccinate synthetase, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Kim, Y, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-23 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Adenylosuccinate Synthetase from
Campylobacter jejuni
To be Published
|
|
3RDW
 
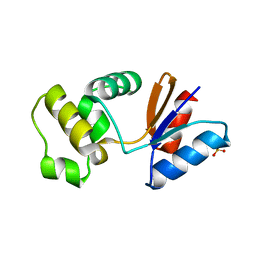 | | Putative arsenate reductase from Yersinia pestis | | Descriptor: | Putative arsenate reductase, SULFATE ION | | Authors: | Osipiuk, J, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-01 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Putative arsenate reductase from Yersinia pestis.
To be Published
|
|
4GFP
 
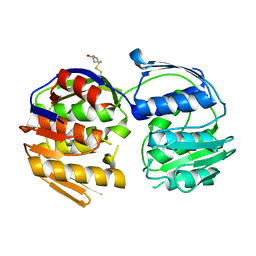 | | 2.7 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in a second conformational state | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, BETA-MERCAPTOETHANOL | | Authors: | Light, S.H, Minasov, G, Krishna, S.N, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-03 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 2.7 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in second conformational state
TO BE PUBLISHED
|
|
4GB7
 
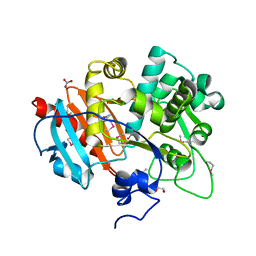 | | Putative 6-aminohexanoate-dimer hydrolase from Bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, 6-aminohexanoate-dimer hydrolase, NITRATE ION | | Authors: | Osipiuk, J, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-07-26 | | Release date: | 2012-08-08 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Putative 6-aminohexanoate-dimer hydrolase from Bacillus anthracis.
To be Published
|
|
4FX5
 
 | | von Willebrand factor type A from Catenulispora acidiphila | | Descriptor: | SODIUM ION, von Willebrand factor type A | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-07-02 | | Release date: | 2012-07-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | von Willebrand factor type A from Catenulispora acidiphila
To be Published
|
|
4FXS
 
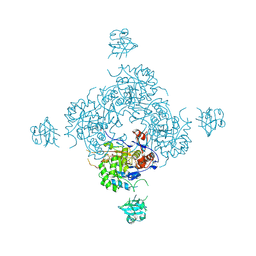 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae complexed with IMP and mycophenolic acid | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, MYCOPHENOLIC ACID, ... | | Authors: | Osipiuk, J, Maltseva, N, Makowska-Grzyska, M, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-07-03 | | Release date: | 2012-07-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae complexed with IMP and mycophenolic acid.
To be Published
|
|
3R2T
 
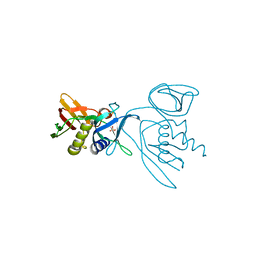 | | 2.2 Angstrom Resolution Crystal Structure of Superantigen-like Protein from Staphylococcus aureus subsp. aureus NCTC 8325. | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Kiryukhina, O, Falugi, F, Bottomley, M, Bagnoli, F, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-14 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 2.2 Angstrom Resolution Crystal Structure of Superantigen-like Protein from Staphylococcus aureus subsp. aureus NCTC 8325.
TO BE PUBLISHED
|
|
4GHJ
 
 | | 1.75 Angstrom Crystal Structure of Transcriptional Regulator ftom Vibrio vulnificus. | | Descriptor: | Probable transcriptional regulator | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 Angstrom Crystal Structure of Transcriptional Regulator ftom Vibrio vulnificus.
TO BE PUBLISHED
|
|
4EBJ
 
 | | Crystal structure of aminoglycoside 4'-O-adenylyltransferase ANT(4')-IIb, apo | | Descriptor: | 1,2-ETHANEDIOL, Aminoglycoside nucleotidyltransferase, SULFATE ION | | Authors: | Stogios, P.J, Wawrzak, Z, Minasov, G, Evdokimova, E, Egorova, O, Yim, V, Kudritska, M, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-23 | | Release date: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of aminoglycoside 4'-O-adenylyltransferase ANT(4')-IIb, apo
To be Published
|
|
4EQB
 
 | | 1.5 Angstrom Crystal Structure of Spermidine/Putrescine ABC Transporter Substrate-Binding Protein PotD from Streptococcus pneumoniae strain Canada MDR_19A in Complex with Calcium and HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Stogios, P.J, Kudritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-18 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom Crystal Structure of Spermidine/Putrescine ABC Transporter Substrate-Binding Protein from Streptococcus pneumoniae strain Canada MDR_19A in Complex with Calcium and HEPES.
TO BE PUBLISHED
|
|
3QTT
 
 | | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis Complexed with Beta-gamma ATP and Beta-alanine | | Descriptor: | BETA-ALANINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-02-23 | | Release date: | 2011-03-23 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis Complexed with Beta-gamma ATP and Beta-alanine.
To be Published
|
|
4E4R
 
 | | EutD phosphotransacetylase from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Zhou, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-13 | | Release date: | 2012-03-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | EutD phosphotransacetylase from Staphylococcus aureus.
To be Published
|
|
4EG9
 
 | | 1.9 Angstrom resolution crystal structure of Se-methionine hypothetical protein SAOUHSC_02783 from Staphylococcus aureus | | Descriptor: | CALCIUM ION, Uncharacterized protein SAOUHSC_02783 | | Authors: | Biancucci, M, Minasov, G, Halavaty, A, Filippova, E.V, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-30 | | Release date: | 2012-04-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom resolution crystal structure of Se-methionine hypothetical protein SAOUHSC_02783 from Staphylococcus aureus
TO BE PUBLISHED
|
|
4EH1
 
 | | Crystal Structure of the Flavohem-like-FAD/NAD Binding Domain of Nitric Oxide Dioxygenase from Vibrio cholerae O1 biovar El Tor | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavohemoprotein, ... | | Authors: | Kim, Y, Gu, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-02 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Flavohem-like-FAD/NAD Binding Domain of Nitric Oxide Dioxygenase from Vibrio cholerae O1 biovar El Tor
To be Published
|
|
3QFG
 
 | | Structure of a putative lipoprotein from Staphylococcus aureus subsp. aureus NCTC 8325 | | Descriptor: | Uncharacterized protein | | Authors: | Filippova, E.V, Halavaty, A, Shuvalova, L, Minasov, G, Dubrovska, I, Winsor, J, Kiryukhina, O, Papazisi, L, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-21 | | Release date: | 2011-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structure of a putative lipoprotein from Staphylococcus aureus subsp. aureus NCTC 8325
To be Published
|
|
4GUI
 
 | | 1.78 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) in Complex with Quinate | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, 3-dehydroquinate dehydratase, NICKEL (II) ION | | Authors: | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structures of type I dehydroquinate dehydratase in complex with quinate and shikimate suggest a novel mechanism of schiff base formation.
Biochemistry, 53, 2014
|
|
4GUF
 
 | | 1.5 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) E86A Mutant | | Descriptor: | 3-dehydroquinate dehydratase, CHLORIDE ION | | Authors: | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Reassessing the type I dehydroquinate dehydratase catalytic triad: Kinetic and structural studies of Glu86 mutants.
Protein Sci., 22, 2013
|
|
3RQT
 
 | | 1.5 Angstrom Crystal Structure of the Complex of Ligand Binding Component of ABC-type Import System from Staphylococcus aureus with Nickel and two Histidines | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HISTIDINE, NICKEL (II) ION, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Kiryukhina, O, Falugi, F, Bottomley, M, Bagnoli, F, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-28 | | Release date: | 2011-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom Crystal Structure of the Complex of Ligand Binding Component of ABC-type Import System from Staphylococcus aureus with Nickel and two Histidines.
TO BE PUBLISHED
|
|
4H3U
 
 | | Crystal structure of hypothetical protein with ketosteroid isomerase-like protein fold from Catenulispora acidiphila DSM 44928 | | Descriptor: | ACETATE ION, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-14 | | Release date: | 2012-10-03 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural characterization of a hypothetical protein: a potential agent involved in trimethylamine metabolism in Catenulispora acidiphila.
J.Struct.Funct.Genom., 15, 2014
|
|
3RF1
 
 | | The crystal structure of glycyl-tRNA synthetase subunit alpha from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | (2S)-2-hydroxybutanedioic acid, GLYCEROL, Glycyl-tRNA synthetase alpha subunit | | Authors: | Tan, K, Zhang, R, Zhou, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-05 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of glycyl-tRNA synthetase subunit alpha from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
3R38
 
 | | 2.23 Angstrom resolution crystal structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase (murA) from Listeria monocytogenes EGD-e | | Descriptor: | CHLORIDE ION, SULFATE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase 1 | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-15 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | 2.23 Angstrom resolution crystal structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase (murA) from Listeria monocytogenes EGD-e
To be Published
|
|
