3BHW
 
 | | Crystal structure of an uncharacterized protein from Magnetospirillum magneticum | | Descriptor: | Uncharacterized protein | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Lau, C, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-29 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an uncharacterized protein from Magnetospirillum magneticum.
To be Published
|
|
3EFK
 
 | | Structure of c-Met with pyrimidone inhibitor 50 | | Descriptor: | 5-{4-[(6,7-dimethoxyquinolin-4-yl)oxy]-3-fluorophenyl}-2-[(4-fluorophenyl)amino]-3-methylpyrimidin-4(3H)-one, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, D'Angelo, N, Whittington, D, Dussault, I. | | Deposit date: | 2008-09-09 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis, and biological evaluation of potent c-Met inhibitors.
J.Med.Chem., 51, 2008
|
|
3BIL
 
 | | Crystal structure of a probable LacI family transcriptional regulator from Corynebacterium glutamicum | | Descriptor: | Probable LacI-family transcriptional regulator | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Mendoza, M, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a probable LacI family transcriptional regulator from Corynebacterium glutamicum.
To be Published
|
|
3BIX
 
 | | Crystal structure of the extracellular esterase domain of Neuroligin-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, NICKEL (II) ION, ... | | Authors: | Arac, D, Boucard, A.A, Ozkan, E, Strop, P, Newell, E, Sudhof, T.C, Brunger, A.T. | | Deposit date: | 2007-12-01 | | Release date: | 2007-12-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Neuroligin-1 and the Neuroligin-1/Neurexin-1beta Complex Reveal Specific Protein-Protein and Protein-Ca(2+) Interactions.
Neuron, 56, 2007
|
|
3B1T
 
 | | Crystal structure of human peptidylarginine deiminase 4 in complex with o-Cl-amidine | | Descriptor: | 2-{[(2S)-1-amino-5-{[(1Z)-2-chloroethanimidoyl]amino}-1-oxopentan-2-yl]carbamoyl}benzoic acid, CALCIUM ION, Protein-arginine deiminase type-4, ... | | Authors: | Causey, C.P, Jones, J.E, Slack, J.L, Kamei, D, Jones Jr, L.E, Subramanian, V, Knuckley, B, Ebrahimi, P, Chumanevich, A.A, Luo, Y, Hashimoto, H, Shimizu, T, Sato, M, Hofseth, L.J, Thompson, P.R. | | Deposit date: | 2011-07-13 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Development of N-alpha-(2-Carboxyl)benzoyl-N(5)-(2-fluoro-1-iminoethyl)-l-ornithine Amide (o-F-amidine) and N-alpha-(2-Carboxyl)benzoyl-N(5)-(2-chloro-1-iminoethyl)-l-ornithine Amide (o-Cl-amidine) As Second Generation Protein Arginine Deiminase (PAD) Inhibitors
J.Med.Chem., 54, 2011
|
|
3U45
 
 | | Human Carbonic Anhydrase II V143A | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, ZINC ION | | Authors: | West, D, Mckenna, R. | | Deposit date: | 2011-10-07 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural Modification of the hydrophobic pocket in Human Carbonic Anhydrase II
To be Published
|
|
3BHJ
 
 | | Crystal structure of human Carbonyl Reductase 1 in complex with glutathione | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 3-(4-AMINO-1-TERT-BUTYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-3-YL)PHENOL, Carbonyl reductase [NADPH] 1, ... | | Authors: | Rauh, D, Bateman, R.L, Shokat, K.M. | | Deposit date: | 2007-11-28 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Human carbonyl reductase 1 is an s-nitrosoglutathione reductase
J.Biol.Chem., 283, 2008
|
|
3EFJ
 
 | | Structure of c-Met with pyrimidone inhibitor 7 | | Descriptor: | 2-benzyl-5-{4-[(6,7-dimethoxyquinolin-4-yl)oxy]-3-fluorophenyl}-3-methylpyrimidin-4(3H)-one, Hepatocyte growth factor receptor | | Authors: | D'Angelo, N, Bellon, S, Whittington, D. | | Deposit date: | 2008-09-09 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis, and biological evaluation of potent c-Met inhibitors.
J.Med.Chem., 51, 2008
|
|
3BGH
 
 | | Crystal structure of putative neuraminyllactose-binding hemagglutinin homolog from Helicobacter pylori | | Descriptor: | Putative neuraminyllactose-binding hemagglutinin homolog, SULFATE ION | | Authors: | Bonanno, J.B, Dickey, J, Bain, K.T, McKenzie, C, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-26 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of putative neuraminyllactose-binding hemagglutinin homolog from Helicobacter pylori.
To be Published
|
|
3E84
 
 | | Structure-function Analysis of 2-Keto-3-deoxy-D-glycero-D-galacto-nononate-9-phosphate (KDN) Phosphatase Defines a New Clad Within the Type C0 HAD Subfamily | | Descriptor: | 1,2-ETHANEDIOL, Acylneuraminate cytidylyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lu, Z, Wang, L, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2008-08-19 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Function Analysis of 2-Keto-3-deoxy-D-glycero-D-galactonononate-9-phosphate Phosphatase Defines Specificity Elements in Type C0 Haloalkanoate Dehalogenase Family Members.
J.Biol.Chem., 284, 2009
|
|
3C1H
 
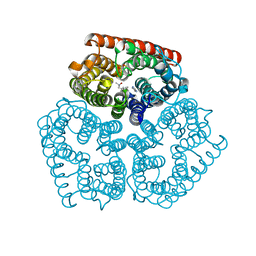 | | Substrate binding, deprotonation and selectivity at the periplasmic entrance of the E. coli ammonia channel AmtB | | Descriptor: | ACETATE ION, Ammonia channel, IMIDAZOLE, ... | | Authors: | Lupo, D, Winkler, F.K. | | Deposit date: | 2008-01-23 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate binding, deprotonation, and selectivity at the periplasmic entrance of the Escherichia coli ammonia channel AmtB.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZNI
 
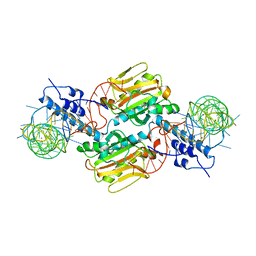 | | Crystal structure of Pyrrolysyl-tRNA synthetase-tRNA(Pyl) complex from Desulfitobacterium hafniense | | Descriptor: | CALCIUM ION, Pyrrolysyl-tRNA synthetase, bacterial tRNA | | Authors: | Nozawa, K, Araiso, Y, Soll, D, Ishitani, R, Nureki, O. | | Deposit date: | 2008-04-25 | | Release date: | 2008-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Pyrrolysyl-tRNA synthetase-tRNA(Pyl) structure reveals the molecular basis of orthogonality
Nature, 457, 2009
|
|
3FZJ
 
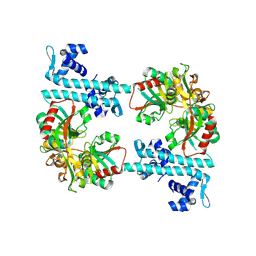 | | TsaR low resolution crystal structure, tetragonal form | | Descriptor: | LysR type regulator of tsaMBCD | | Authors: | Monferrer, D, Tralau, T, Kertesz, M.A, Dix, I, Kikhney, A.G, Svergun, D.I, Uson, I. | | Deposit date: | 2009-01-26 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (7.1 Å) | | Cite: | Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold
Mol.Microbiol., 75, 2010
|
|
2ZT9
 
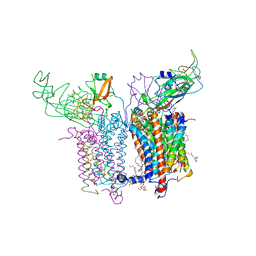 | | Crystal Structure of the Cytochrome b6f Complex from Nostoc sp. PCC 7120 | | Descriptor: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, Apocytochrome f, ... | | Authors: | Craner, W.A, Baniulis, D, Yamashita, E. | | Deposit date: | 2008-09-27 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Function, Stability, and Chemical Modification of the Cyanobacterial Cytochrome b6f Complex from Nostoc sp. PCC 7120
J.Biol.Chem., 284, 2009
|
|
3G3V
 
 | | Crystal structure of spin labeled T4 Lysozyme (V131R1) at 291 K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Hubbell, W.L. | | Deposit date: | 2009-02-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins.
Protein Sci., 18, 2009
|
|
1FVO
 
 | | CRYSTAL STRUCTURE OF HUMAN ORNITHINE TRANSCARBAMYLASE COMPLEXED WITH CARBAMOYL PHOSPHATE | | Descriptor: | ORNITHINE TRANSCARBAMYLASE, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Shi, D, Morizono, H, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2000-09-20 | | Release date: | 2001-04-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human ornithine transcarbamylase: crystallographic insights into substrate recognition and conformational changes.
Biochem.J., 354, 2001
|
|
4G3H
 
 | | Crystal structure of helicobacter pylori arginase | | Descriptor: | Arginase (RocF), MANGANESE (II) ION | | Authors: | Zhang, J, Zhang, X, Li, D, Hu, Y, Zou, Q, Wang, D. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function studies on Helicobacter pylori arginase
To be Published
|
|
1UY0
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with glc-1,3-glc-1,4-glc-1,3-glc | | Descriptor: | CALCIUM ION, CELLULASE B, CHLORIDE ION, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
3G3W
 
 | | Crystal structure of spin labeled T4 Lysozyme (T151R1) at 291 K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Hubbell, W.L. | | Deposit date: | 2009-02-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins.
Protein Sci., 18, 2009
|
|
2Z35
 
 | | Crystal structure of immune receptor | | Descriptor: | T-cell receptor alpha-chain, T-cell receptor beta-chain | | Authors: | Feng, D, Bond, C.J, Ely, L.K, Garcia, K.C. | | Deposit date: | 2007-06-01 | | Release date: | 2007-10-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction 'codon'
Nat.Immunol., 8, 2007
|
|
2ZNJ
 
 | | Crystal structure of Pyrrolysyl-tRNA synthetase from Desulfitobacterium hafniense | | Descriptor: | Putative uncharacterized protein | | Authors: | Nozawa, K, Araiso, Y, Soll, D, Ishitani, R, Nureki, O. | | Deposit date: | 2008-04-25 | | Release date: | 2008-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pyrrolysyl-tRNA synthetase-tRNA(Pyl) structure reveals the molecular basis of orthogonality
Nature, 457, 2009
|
|
2Z7J
 
 | | Structural insights into de multifunctional VP3 protein of birnaviruses:gold derivative | | Descriptor: | Capsid assembly protein VP3, GOLD ION | | Authors: | Casanas, A, Navarro, A, Ferrer-Orta, C, Gonzalez, D, Rodriguez, J.F, Verdaguer, N. | | Deposit date: | 2007-08-23 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the multifunctional protein VP3 of birnaviruses.
Structure, 16, 2008
|
|
3GDG
 
 | | Crystal structure of the NADP-dependent mannitol dehydrogenase from Cladosporium herbarum. | | Descriptor: | Probable NADP-dependent mannitol dehydrogenase, SODIUM ION | | Authors: | Nuess, D, Goettig, P, Magler, I, Denk, U, Breitenbach, M, Schneider, P.B, Brandstetter, H, Simon-Nobbe, B. | | Deposit date: | 2009-02-24 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the NADP-dependent mannitol dehydrogenase from Cladosporium herbarum: Implications for oligomerisation and catalysis.
Biochimie, 92, 2010
|
|
2Z90
 
 | | Crystal Structure of the Second Dps from Mycobacterium smegmatis | | Descriptor: | CHLORIDE ION, FE (II) ION, MAGNESIUM ION, ... | | Authors: | Roy, S, Saraswathi, R, Chatterji, D, Vijayan, M. | | Deposit date: | 2007-09-13 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies on the second Mycobacterium smegmatis Dps: invariant and variable features of structure, assembly and function.
J.Mol.Biol., 375, 2008
|
|
3BKC
 
 | | Crystal structure of anti-amyloid beta FAB WO2 (P21, FormB) | | Descriptor: | SODIUM ION, WO2 IgG2a Fab fragment Heavy Chain, WO2 IgG2a Fab fragment Light Chain Kappa | | Authors: | Miles, L.A, Wun, K.S, Crespi, G.A, Fodero-Tavoletti, M, Galatis, D, Bageley, C.J, Beyreuther, K, Masters, C.L, Cappai, R, McKinstry, W.J, Barnham, K.J, Parker, M.W. | | Deposit date: | 2007-12-06 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope.
J.Mol.Biol., 377, 2008
|
|
