6JKY
 
 | |
7Q8B
 
 | | Leishmania major actin filament in ADP-Pi state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, MAGNESIUM ION, ... | | Authors: | Kotila, T, Muniyandi, S, Lappalainen, P, Huiskonen, J.T. | | Deposit date: | 2021-11-11 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of rapid actin dynamics in the evolutionarily divergent Leishmania parasite.
Nat Commun, 13, 2022
|
|
7Q8C
 
 | | Leishmania major actin filament in ADP-state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, MAGNESIUM ION | | Authors: | Kotila, T, Muniyandi, S, Lappalainen, P, Huiskonen, J.T. | | Deposit date: | 2021-11-11 | | Release date: | 2022-05-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural basis of rapid actin dynamics in the evolutionarily divergent Leishmania parasite.
Nat Commun, 13, 2022
|
|
7Q8S
 
 | | Leishmania major ADP-actin filament decorated with Leishmania major cofilin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADF/Cofilin, Actin, ... | | Authors: | Kotila, T, Muniyandi, S, Lappalainen, P, Huiskonen, J.T. | | Deposit date: | 2021-11-11 | | Release date: | 2022-05-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of rapid actin dynamics in the evolutionarily divergent Leishmania parasite.
Nat Commun, 13, 2022
|
|
7TFF
 
 | | Crystal structure of human platelet phosphofructokinase-1 mutant- D564N | | Descriptor: | ATP-dependent 6-phosphofructokinase, platelet type, PHOSPHATE ION, ... | | Authors: | Hansen, H, Webb, B.A, Robart, A.R, Narayanasami, S. | | Deposit date: | 2022-01-06 | | Release date: | 2023-07-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Cancer-associated somatic mutations in human phosphofructokinase-1 reveal a critical electrostatic interaction for allosteric regulation of enzyme activity.
Biochem.J., 480, 2023
|
|
8FA3
 
 | |
6K4L
 
 | |
6IST
 
 | | Crystal structure of a wild type endolysin LysIME-EF1 | | Descriptor: | CALCIUM ION, Lysin | | Authors: | Ouyang, S.Y. | | Deposit date: | 2018-11-19 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and functional insights into a novel two-component endolysin encoded by a single gene in Enterococcus faecalis phage.
Plos Pathog., 16, 2020
|
|
6IM4
 
 | |
6K4R
 
 | |
6K4K
 
 | |
6KSF
 
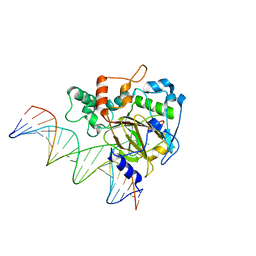 | | Crystal Structure of ALKBH1 bound to 21-mer DNA bulge | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 1, CHLORIDE ION, DNA (5'-D(*DGP*DCP*DTP*DGP*DAP*DGP*DTP*DGP*DCP*DCP*DCP*DGP*DCP*DGP*DTP*DGP*DCP*DTP*DGP*DGP*DAP*DTP*DCP*DC)-3'), ... | | Authors: | Li, H, Zhang, M. | | Deposit date: | 2019-08-23 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
6KS5
 
 | |
2I8E
 
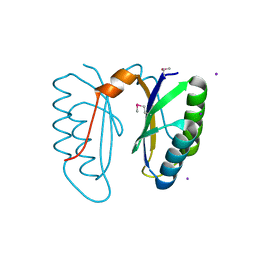 | | Structure of SSO1404, a predicted DNA repair-associated protein from Sulfolobus solfataricus P2 | | Descriptor: | Hypothetical protein, IODIDE ION | | Authors: | Wang, S, Zimmerman, M.D, Kudritska, M, Chruszcz, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-01 | | Release date: | 2006-09-26 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats.
J.Biol.Chem., 283, 2008
|
|
3TJH
 
 | | 42F3-p3A1/H2-Ld complex | | Descriptor: | 42F3 alpha, 42F3 beta, H2-Ld SBM2, ... | | Authors: | Adams, J.J, Kruse, A, Kranz, D.M, Garcia, K.C. | | Deposit date: | 2011-08-24 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex.
Immunity, 35, 2011
|
|
3SVU
 
 | |
3SVR
 
 | |
3SVO
 
 | |
3SVS
 
 | |
2VP1
 
 | | Fe-FutA2 from Synechocystis PCC6803 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FE (III) ION, ... | | Authors: | Badarau, A, Firbank, S.J, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Futa2 is a Ferric Binding Protein from Synechocystis Pcc 6803.
J.Biol.Chem., 283, 2008
|
|
3SVN
 
 | |
1AMX
 
 | |
3TFK
 
 | | 42F3-p4B10/H2-Ld | | Descriptor: | 42F3 alpha, 42F3 beta, H2-Ld SBM2, ... | | Authors: | Adams, J.J, Kranz, D.M, Garcia, K.C. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex.
Immunity, 35, 2011
|
|
3TF7
 
 | | 42F3 QL9/H2-Ld complex | | Descriptor: | 42F3 Mut7 scFv (42F3 alpha chain, linker, 42F3 beta chain), ... | | Authors: | Adams, J.J, Kranz, D.M, Garcia, K.C. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex.
Immunity, 35, 2011
|
|
3TPU
 
 | | 42F3 p5E8/H2-Ld complex | | Descriptor: | 1,2-ETHANEDIOL, 42F3 alpha, 42F3 beta, ... | | Authors: | Adams, J.J, Kranz, D.M, Garcia, K.C. | | Deposit date: | 2011-09-08 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex.
Immunity, 35, 2011
|
|
