5X58
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5B
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 2 | | Descriptor: | Spike glycoprotein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X59
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, three-fold symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
2QJT
 
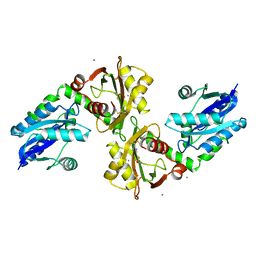 | | Crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase complexed with AMP and MN ion from Francisella tularensis | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Raffaelli, N, Magni, G, Grishin, N.V, Osterman, A, Zhang, H. | | Deposit date: | 2007-07-09 | | Release date: | 2008-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
5C98
 
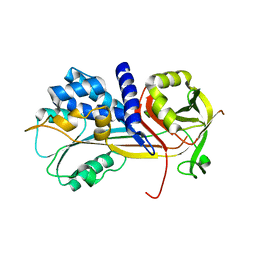 | | 1.45A resolution structure of SRPN18 from Anopheles gambiae | | Descriptor: | AGAP007691-PB | | Authors: | Lovell, S, Battaile, K.P, Gulley, M, Zhang, X, Meekins, D.A, Gao, F.P, Michel, K. | | Deposit date: | 2015-06-26 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 angstrom resolution structure of SRPN18 from the malaria vector Anopheles gambiae.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
5X5C
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 1 | | Descriptor: | S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
2LW6
 
 | | Solution structure of an avirulence protein AvrPiz-t from pathogen Magnaportheoryzae | | Descriptor: | AvrPiz-t protein | | Authors: | Zhang, Z.-M, Zhang, X, Zhou, Z, Hu, H, Liu, M, Zhou, B, Zhou, J. | | Deposit date: | 2012-07-23 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Magnaporthe oryzae avirulence protein AvrPiz-t.
J.Biomol.Nmr, 55, 2013
|
|
8WTM
 
 | | Cryo-EM structure of jasmonic acid transporter ABCG16 bound to JA | | Descriptor: | ABC transporter G family member 16, {(1R,2R)-3-oxo-2-[(2Z)-pent-2-en-1-yl]cyclopentyl}acetic acid | | Authors: | Huang, X, An, N, Zhang, X, Zhang, P. | | Deposit date: | 2023-10-19 | | Release date: | 2024-10-23 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Cryo-EM structure and molecular mechanism of the jasmonic acid transporter ABCG16.
Nat.Plants, 10, 2024
|
|
8WTO
 
 | | Cryo-EM structure of jasmonic acid transporter ABCG16 in outward conformation | | Descriptor: | ABC transporter G family member 16, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Huang, X, An, N, Zhang, X, Zhang, P. | | Deposit date: | 2023-10-19 | | Release date: | 2024-10-23 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM structure and molecular mechanism of the jasmonic acid transporter ABCG16.
Nat.Plants, 10, 2024
|
|
8WTP
 
 | |
8WTN
 
 | | Cryo-EM structure of jasmonic acid transporter ABCG16 in occluded conformation | | Descriptor: | ABC transporter G family member 16, ADENOSINE-5'-DIPHOSPHATE, VANADATE ION | | Authors: | Huang, X, An, N, Zhang, X, Zhang, P. | | Deposit date: | 2023-10-19 | | Release date: | 2024-10-23 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-EM structure and molecular mechanism of the jasmonic acid transporter ABCG16.
Nat.Plants, 10, 2024
|
|
4EW0
 
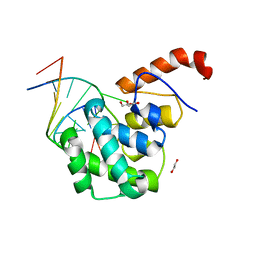 | | mouse MBD4 glycosylase domain in complex with a G:5hmU (5-hydroxymethyluracil) mismatch | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*GP*(5HU)P*GP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2012-04-26 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
1VAZ
 
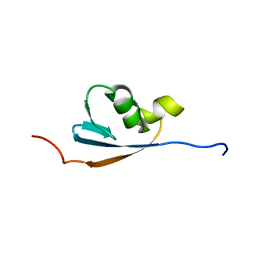 | | Solution structures of the p47 SEP domain | | Descriptor: | NSFL1 cofactor p47 | | Authors: | Yuan, X, Simpson, P, Mckeown, C, Kondo, H, Uchiyama, K, Wallis, R, Dreveny, I, Keetch, C, Zhang, X, Robinson, C, Freemont, P, Matthews, S. | | Deposit date: | 2004-02-20 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and interactions of p47, a major adaptor of the AAA ATPase, p97.
Embo J., 23, 2004
|
|
4M8N
 
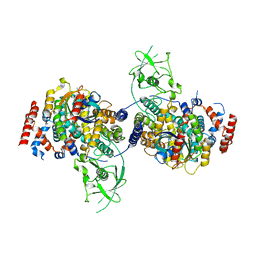 | | Crystal Structure of PlexinC1/Rap1B Complex | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pascoe, H.G, Wang, Y, Brautigam, C.A, He, H, Zhang, X. | | Deposit date: | 2013-08-13 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.294 Å) | | Cite: | Structural basis for activation and non-canonical catalysis of the Rap GTPase activating protein domain of plexin.
Elife, 2, 2013
|
|
4EVV
 
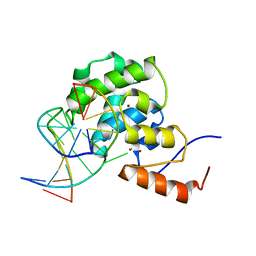 | | mouse MBD4 glycosylase domain in complex with a G:T mismatch | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*GP*TP*GP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2012-04-26 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
4EW4
 
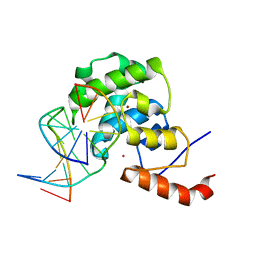 | | mouse MBD4 glycosylase domain in complex with DNA containing a ribose sugar | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*GP*(3DR)P*GP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), Methyl-CpG-binding domain protein 4, ... | | Authors: | Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2012-04-26 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.791 Å) | | Cite: | Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
4F0Q
 
 | | MspJI Restriction Endonuclease - P21 Form | | Descriptor: | MAGNESIUM ION, Restriction endonuclease | | Authors: | Horton, J.R, Mabuchi, M, Cohen-Karni, D, Zhang, X, Griggs, R, Samaranayake, M, Roberts, R.J, Zheng, Y, Cheng, X. | | Deposit date: | 2012-05-04 | | Release date: | 2012-08-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.046 Å) | | Cite: | Structure and cleavage activity of the tetrameric MspJI DNA modification-dependent restriction endonuclease.
Nucleic Acids Res., 40, 2012
|
|
4F0P
 
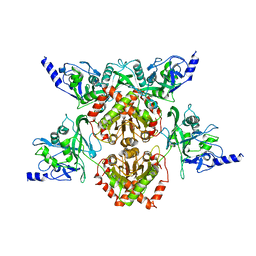 | | MspJI Restriction Endonuclease - P31 Form | | Descriptor: | MAGNESIUM ION, Restriction endonuclease | | Authors: | Horton, J.R, Mabuchi, M, Cohen-Karni, D, Zhang, X, Griggs, R, Samaranayake, M, Roberts, R.J, Zheng, Y, Cheng, X. | | Deposit date: | 2012-05-04 | | Release date: | 2012-08-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure and cleavage activity of the tetrameric MspJI DNA modification-dependent restriction endonuclease.
Nucleic Acids Res., 40, 2012
|
|
5ABH
 
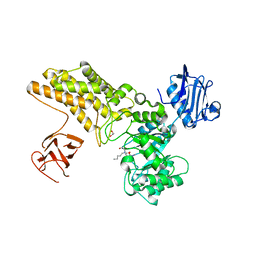 | | Structure of GH84 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2R,3S,4R,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)-1-pentyl-pyrrolidin-2-yl]-N-methyl-ethanamide, CALCIUM ION, ... | | Authors: | Bergeron-Brlek, M, Goodwin-Tindall, J, Cekic, N, Varghese, V, Zandberg, W.F, Shan, X, Roth, C, Chan, S, Davies, G.J, Vocadlo, D.J, Britton, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Convenient Approach to Stereoisomeric Iminocyclitols: Generation of Potent Brain-Permeable Oga Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7XY4
 
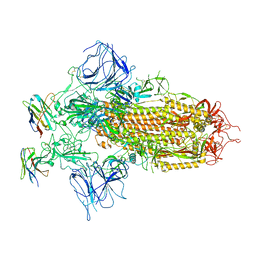 | |
7XY3
 
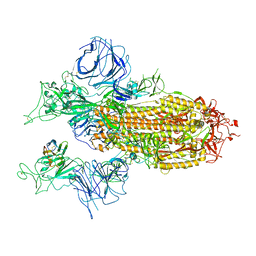 | |
8KD3
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification, H3K9Q mutation and 187bp DNA | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD4
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class1 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
8KD6
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class3 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
5ABG
 
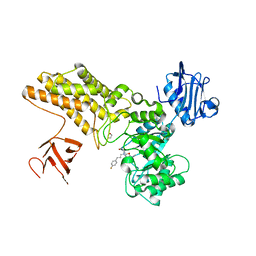 | | Structure of GH84 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2R,3S,4R,5R)-1-[3-(4-fluorophenyl)propyl]-5-(hydroxymethyl)-3,4-bis(oxidanyl)pyrrolidin-2-yl]-N-methyl-ethanamide, CALCIUM ION, ... | | Authors: | Bergeron-Brlek, M, Goodwin-Tindall, J, Cekic, N, Varghese, V, Zandberg, W.F, Shan, X, Roth, C, Chan, S, Davies, G.J, Vocadlo, D.J, Britton, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Convenient Approach to Stereoisomeric Iminocyclitols: Generation of Potent Brain-Permeable Oga Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
