6OA9
 
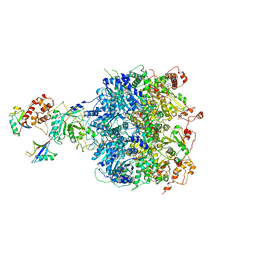 | | Cdc48-Npl4 complex processing poly-ubiquitinated substrate in the presence of ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 48, ... | | Authors: | Twomey, E.C, Ji, Z, Wales, T.E, Bodnar, N.O, Engen, J.R, Rapoport, T.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding.
Science, 365, 2019
|
|
5ES1
 
 | |
2Z4B
 
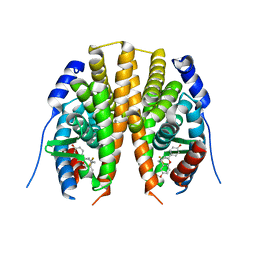 | | Estrogen receptor beta ligand-binding domain complexed to a benzopyran ligand | | Descriptor: | (3AS,4R,9BR)-2,2-DIFLUORO-4-(4-HYDROXYPHENYL)-1,2,3,3A,4,9B-HEXAHYDROCYCLOPENTA[C]CHROMEN-8-OL, Estrogen receptor beta | | Authors: | Wang, Y. | | Deposit date: | 2007-06-14 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Benzopyrans as selective estrogen receptor beta agonists (SERBAs). Part 3: Synthesis of cyclopentanone and cyclohexanone intermediates for C-ring modification.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5F16
 
 | | CTA-modified hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | McGlone, C, Nix, J.C, Page, R.C. | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Investigating the Impact of Polymer Functional Groups on the Stability and Activity of Lysozyme-Polymer Conjugates.
Biomacromolecules, 17, 2016
|
|
6OBZ
 
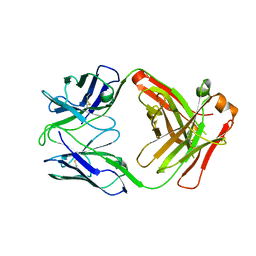 | | Crystal structure of FluA-20 Fab | | Descriptor: | Heavy chain of FluA-20 Fab, Light chain of FluA-20 Fab, Immunoglobulin light chain | | Authors: | Wilson, I.A, Lang, S. | | Deposit date: | 2019-03-21 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A Site of Vulnerability on the Influenza Virus Hemagglutinin Head Domain Trimer Interface.
Cell, 177, 2019
|
|
6OIT
 
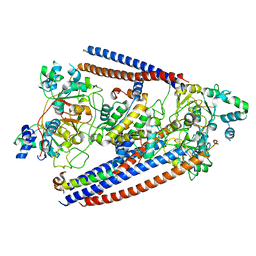 | | CryoEM structure of Arabidopsis DDR' complex (DRD1 peptide-DMS3-RDM1) | | Descriptor: | Protein CHROMATIN REMODELING 35, Protein DEFECTIVE IN MERISTEM SILENCING 3, Protein RDM1 | | Authors: | Wongpalee, S.P, Liu, S, Zhou, Z.H, Jacobsen, S.E. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | CryoEM structures of Arabidopsis DDR complexes involved in RNA-directed DNA methylation.
Nat Commun, 10, 2019
|
|
6OKZ
 
 | |
6ONP
 
 | |
6ONQ
 
 | |
2CD8
 
 | | Crystal structure of YC-17-bound cytochrome P450 PikC (CYP107L1) | | Descriptor: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yermalitskaya, L.I, Kim, Y, Sherman, D.H, Waterman, M.R, Podust, L.M. | | Deposit date: | 2006-01-20 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis for Substrate Anchoring, Active Site Selectivity, and Product Formation by P450 Pikc from Streptomyces Venezuelae.
J.Biol.Chem., 281, 2006
|
|
6OT9
 
 | | Bimetallic dodecameric cage design 1 (BMC1) from cytochrome cb562 | | Descriptor: | ACETOHYDROXAMIC ACID, FE (III) ION, HEME C, ... | | Authors: | Golub, E, Esselborn, J, Bailey, J.B, Tezcan, F.A. | | Deposit date: | 2019-05-02 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Constructing protein polyhedra via orthogonal chemical interactions.
Nature, 578, 2020
|
|
3AVH
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
5FP3
 
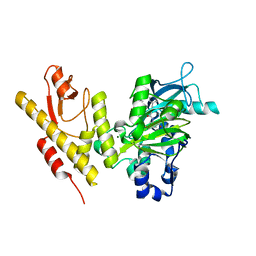 | |
3AV9
 
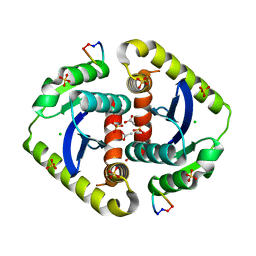 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-02 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3AVF
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3AVN
 
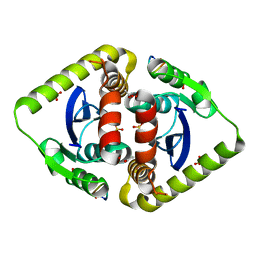 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
6OT4
 
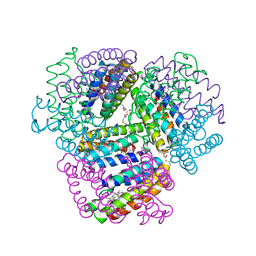 | | Bimetallic dodecameric cage design 2 (BMC2) from cytochrome cb562 | | Descriptor: | ACETOHYDROXAMIC ACID, FE (III) ION, HEME C, ... | | Authors: | Golub, E, Esselborn, J, Bailey, J.B, Tezcan, F.A. | | Deposit date: | 2019-05-02 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Constructing protein polyhedra via orthogonal chemical interactions.
Nature, 578, 2020
|
|
3AVL
 
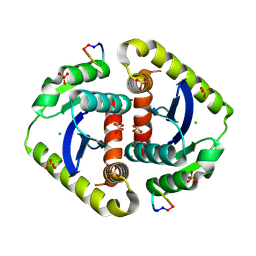 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
6OT7
 
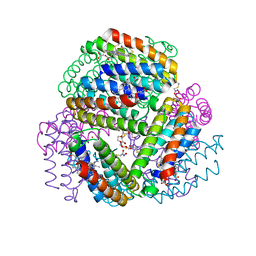 | | Bimetallic dodecameric cage design 3 (BMC3) from cytochrome cb562 | | Descriptor: | ACETOHYDROXAMIC ACID, FE (III) ION, HEME C, ... | | Authors: | Golub, E, Esselborn, J, Bailey, J.B, Tezcan, F.A. | | Deposit date: | 2019-05-02 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Constructing protein polyhedra via orthogonal chemical interactions.
Nature, 578, 2020
|
|
6OT8
 
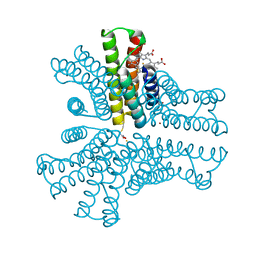 | | Bimetallic hexameric cage design 4 (BMC4) from cytochrome cb562 | | Descriptor: | ACETOHYDROXAMIC ACID, FE (III) ION, HEME C, ... | | Authors: | Golub, E, Esselborn, J, Bailey, J.B, Tezcan, F.A. | | Deposit date: | 2019-05-02 | | Release date: | 2020-01-29 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Constructing protein polyhedra via orthogonal chemical interactions.
Nature, 578, 2020
|
|
5FP4
 
 | |
3AVA
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-02 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3AVI
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-02-15 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3AVB
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-02 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
3AVJ
 
 | | Crystal structures of novel allosteric peptide inhibitors of HIV integrase in the LEDGF binding site | | Descriptor: | ACETIC ACID, CHLORIDE ION, Integrase, ... | | Authors: | Peat, T.S, Deadman, J.J, Newman, J, Rhodes, D.I. | | Deposit date: | 2011-03-05 | | Release date: | 2012-01-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site.
Chembiochem, 12, 2011
|
|
