3BOV
 
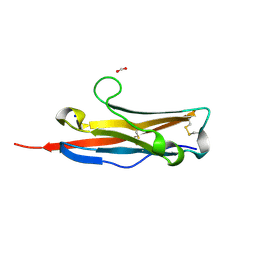 | | Crystal structure of the receptor binding domain of mouse PD-L2 | | Descriptor: | FORMIC ACID, Programmed cell death 1 ligand 2, SODIUM ION | | Authors: | Lazar-Molnar, E, Ramagopal, U, Cao, E, Toro, R, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-12-17 | | Release date: | 2008-07-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of the complex between programmed death-1 (PD-1) and its ligand PD-L2.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3BY5
 
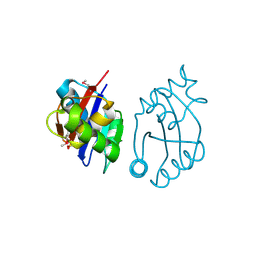 | | Crystal structure of cobalamin biosynthesis protein chiG from Agrobacterium tumefaciens str. C58 | | Descriptor: | Cobalamin biosynthesis protein, SULFATE ION | | Authors: | Patskovsky, Y, Bonanno, J.B, Sojitra, S, Rutter, M, Iizuka, M, Maletic, M, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-15 | | Release date: | 2008-01-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of cobalamin biosynthesis protein from Agrobacterium tumefaciens str. C58.
To be Published
|
|
3C3M
 
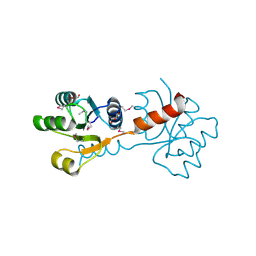 | | Crystal structure of the N-terminal domain of response regulator receiver protein from Methanoculleus marisnigri JR1 | | Descriptor: | GLYCEROL, Response regulator receiver protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Dickey, M, Chang, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the N-terminal domain of response regulator receiver protein from Methanoculleus marisnigri JR1.
To be Published
|
|
3BMA
 
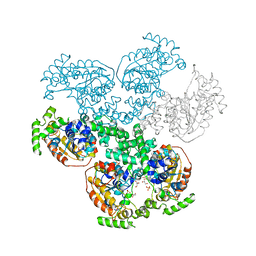 | | Crystal structure of D-alanyl-lipoteichoic acid synthetase from Streptococcus pneumoniae R6 | | Descriptor: | D-alanyl-lipoteichoic acid synthetase, GLYCEROL, SULFATE ION | | Authors: | Patskovsky, Y, Sridhar, V, Bonanno, J.B, Smith, D, Rutter, M, Iizuka, M, Koss, J, Bain, K, Gheyi, T, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-12 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of probable D-Alanyl-Lipoteichoic Acid Synthetase from Streptococcus pneumoniae.
To be Published
|
|
3CS2
 
 | | Crystal structure of PTE G60A mutant | | Descriptor: | CACODYLATE ION, COBALT (II) ION, Parathion hydrolase | | Authors: | Kim, J, Almo, S.C. | | Deposit date: | 2008-04-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of diethyl phosphate bound to the binuclear metal center of phosphotriesterase.
Biochemistry, 47, 2008
|
|
3CS3
 
 | | Crystal structure of sugar-binding transcriptional regulator (LacI family) from Enterococcus faecalis | | Descriptor: | GLYCEROL, SULFATE ION, Sugar-binding transcriptional regulator, ... | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Iizuka, M, Groshong, C, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-08 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of sugar-binding transcriptional regulator (LacI family) from Enterococcus faecalis.
To be Published
|
|
2ZAY
 
 | | Crystal structure of response regulator from Desulfuromonas acetoxidans | | Descriptor: | Response regulator receiver protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-12 | | Release date: | 2007-10-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of response regulator from Desulfuromonas acetoxidans.
To be Published
|
|
3CYM
 
 | | Crystal structure of protein BAD_0989 from Bifidobacterium adolescentis | | Descriptor: | GLYCEROL, SODIUM ION, Uncharacterized protein BAD_0989 | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Chang, S, Bain, K, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of protein BAD_0989 from Bifidobacterium adolescentis.
To be Published
|
|
3CZ8
 
 | | Crystal structure of putative sporulation-specific glycosylase ydhD from Bacillus subtilis | | Descriptor: | GLYCEROL, Putative sporulation-specific glycosylase ydhD | | Authors: | Patskovsky, Y, Romero, R, Rutter, M, Chang, S, Maletic, M, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-28 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative glycosylase ydhD from Bacillus subtilis.
To be Published
|
|
3D47
 
 | | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg and D-malate | | Descriptor: | D-MALATE, MAGNESIUM ION, Putative galactonate dehydratase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-05-14 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg and D-malate.
To be Published
|
|
3CFY
 
 | | Crystal structure of signal receiver domain of putative Luxo repressor protein from Vibrio parahaemolyticus | | Descriptor: | Putative LuxO repressor protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Fong, R, Freeman, J, Iizuka, M, Groshong, C, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Putative Luxo Repressor Protein from Vibrio Parahaemolyticus.
To be Published
|
|
3CG4
 
 | | Crystal structure of response regulator receiver domain protein (CheY-like) from Methanospirillum hungatei JF-1 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Response regulator receiver domain protein (CheY-like) | | Authors: | Patskovsky, Y, Freeman, J, Hu, S, Bain, K, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of Response Regulator Receiver Domain Protein (Chey-Like) from Methanospirillum hungatei JF-1.
To be Published
|
|
3CLW
 
 | | Crystal structure of conserved exported protein from Bacteroides fragilis | | Descriptor: | Conserved exported protein | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Chang, S, Ozyurt, K, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-20 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Crystal structure of conserved exported protein from Bacteroides fragilis.
To be Published
|
|
3CXO
 
 | | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg and 3-deoxy-L-rhamnonate | | Descriptor: | (2R,4S)-2,4,7-trihydroxyheptanoic acid, 3,6-dideoxy-L-arabino-hexonic acid, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Hubbard, B.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-04-24 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: L-rhamnonate dehydratase.
Biochemistry, 47, 2008
|
|
3D19
 
 | | Crystal structure of a conserved metalloprotein from Bacillus cereus | | Descriptor: | Conserved metalloprotein, FE (III) ION, MAGNESIUM ION | | Authors: | Bonanno, J.B, Patskovsky, Y, Freeman, J, Bain, K.T, Chang, S, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-05-05 | | Release date: | 2008-07-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a conserved metalloprotein from Bacillus cereus.
To be Published
|
|
3CZB
 
 | | Crystal structure of putative transglycosylase from Caulobacter crescentus | | Descriptor: | Putative transglycosylase, SULFATE ION | | Authors: | Ramagopal, U.A, Chattopadhyay, K, Toro, R, Wasserman, S, Freeman, J, Logan, C, Bain, K, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-28 | | Release date: | 2008-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of putative transglycosylase from Caulobacter crescentus.
To be Published
|
|
3D46
 
 | | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg and L-tartrate | | Descriptor: | L(+)-TARTARIC ACID, MAGNESIUM ION, Putative galactonate dehydratase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-05-14 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of L-rhamnonate dehydratase from Salmonella typhimurium complexed with Mg and L-tartrate.
To be Published
|
|
3CNB
 
 | | Crystal structure of signal receiver domain of DNA binding response regulator protein (merR) from Colwellia psychrerythraea 34H | | Descriptor: | DNA-binding response regulator, merR family | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Hu, S, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of signal receiver domain of DNA binding response regulator (merR) from Colwellia psychrerythraea 34H.
To be Published
|
|
3CWV
 
 | | Crystal structure of B-subunit of the DNA gyrase from Myxococcus xanthus | | Descriptor: | DNA gyrase, B subunit, truncated | | Authors: | Ramagopal, U.A, Toro, R, Meyer, A.J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-22 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of B-subunit of the DNA gyrase from Myxococcus xanthus.
To be published
|
|
4PGP
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM DESULFOVIBRIO ALASKENSIS G20 (Dde_0634, TARGET EFI-510120) WITH BOUND 3-INDOLE ACETIC ACID | | Descriptor: | 1,2-ETHANEDIOL, 1H-INDOL-3-YLACETIC ACID, Extracellular solute-binding protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-05-02 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PMJ
 
 | | Crystal structure of a putative oxidoreductase from Sinorhizobium meliloti 1021 in complex with NADP | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative oxidoreductase | | Authors: | Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Szlachta, K, Zimmerman, M.D, Hillerich, B.S, Gizzi, A, Toro, R, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-05-21 | | Release date: | 2014-06-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a putative oxidoreductase from Sinorhizobiummeliloti 1021 in complex with NADP
to be published
|
|
1Q6W
 
 | | X-Ray structure of Monoamine oxidase regulatory protein from Archaeoglobus fulgius | | Descriptor: | monoamine oxidase regulatory protein, putative | | Authors: | Fedorov, A.A, Fedorov, E.V, Thirumuruhan, R, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-08-14 | | Release date: | 2003-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | X-ray structure of monoamine oxidase regulatory protein from Archaeoglobus fulgidus
To be Published
|
|
4PX1
 
 | | CRYSTAL STRUCTURE OF Maleylacetoacetate isomerase from Methylobacteriu extorquens AM1 WITH BOUND MALONATE (TARGET EFI-507068) | | Descriptor: | CHLORIDE ION, MALONIC ACID, Maleylacetoacetate isomerase (Glutathione S-transferase) | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of glutathione s-transferase zeta from methylobacterium extorquens (TARGET EFI-507068)
To be Published
|
|
4PXO
 
 | | Crystal structure of Maleylacetoacetate isomerase from Methylobacteriu extorquens AM1 WITH BOUND MALONATE AND GSH (TARGET EFI-507068) | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, MALONIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-24 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of glutathione s-transferase zeta from Methylobacterium extorquens (TARGET EFI-507068)
To be Published
|
|
1RNW
 
 | | RECOMBINANT RIBONUCLEASE A CRYSTALLIZED FROM 80% AMMONIUM SULPHATE | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Fedorov, A.A, Joseph-Mccarthy, D, Fedorov, E.V, Sirakova, D, Graf, I, Almo, S.C. | | Deposit date: | 1996-11-08 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ionic interactions in crystalline bovine pancreatic ribonuclease A.
Biochemistry, 35, 1996
|
|
