8JZE
 
 | | PSI-AcpPCI supercomplex from Symbiodinium | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Li, Z.H, Li, X.Y, Wang, W.D. | | Deposit date: | 2023-07-05 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structures and organizations of PSI-AcpPCI supercomplexes from red tidal and coral symbiotic photosynthetic dinoflagellates.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JW0
 
 | | PSI-AcpPCI supercomplex from Amphidinium carterae | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Li, Z.H, Li, X.Y, Wang, W.D. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and organizations of PSI-AcpPCI supercomplexes from red tidal and coral symbiotic photosynthetic dinoflagellates.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JZF
 
 | | PSI-AcpPCI supercomplex from Symbiodinium | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Li, X.Y, Li, Z.H, Wang, W.D. | | Deposit date: | 2023-07-05 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures and organizations of PSI-AcpPCI supercomplexes from red tidal and coral symbiotic photosynthetic dinoflagellates.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5UR1
 
 | | FGFR1 kinase domain complex with SN37333 in reversible binding mode | | Descriptor: | 3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-{1-[4-(dimethylamino)but-2-enoyl]piperidin-4-yl}-7-(phenylamino)-3,4-dihydropyrimido[4,5-d]pyrimidin-2(1H)-one, Fibroblast growth factor receptor 1 | | Authors: | Yosaatmadja, Y, Paik, W.-K, Smaill, J.B, Squire, C.J. | | Deposit date: | 2017-02-09 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2-Oxo-3, 4-dihydropyrimido[4, 5-d]pyrimidinyl derivatives as new irreversible pan fibroblast growth factor receptor (FGFR) inhibitors.
Eur J Med Chem, 135, 2017
|
|
6IRL
 
 | |
8WGH
 
 | | Cryo-EM structure of the red-shifted Fittonia albivenis PSI-LHCI | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Huang, G.Q, Li, X.X, Sui, S.F, Qin, X.C. | | Deposit date: | 2023-09-21 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure of the red-shifted Fittonia albivenis photosystem I.
Nat Commun, 15, 2024
|
|
8JA0
 
 | | Cryo-EM structure of the NmeCas9-sgRNA-AcrIIC4 ternary complex | | Descriptor: | CRISPR-associated endonuclease Cas9, RNA (117-MER), Uncharacterized protein | | Authors: | Yin, H, Li, Z, Yu, G.M, Li, X.Z. | | Deposit date: | 2023-05-05 | | Release date: | 2023-08-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Inhibitory mechanism of CRISPR-Cas9 by AcrIIC4.
Nucleic Acids Res., 51, 2023
|
|
8VH5
 
 | | Cryo-EM structure of Rab12-LRRK2 complex in the LRRK2 dimer state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2, MAGNESIUM ION, ... | | Authors: | Zhu, H, Sun, J. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-09 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | RAB12-LRRK2 complex suppresses primary ciliogenesis and regulates centrosome homeostasis in astrocytes.
Nat Commun, 15, 2024
|
|
8VH4
 
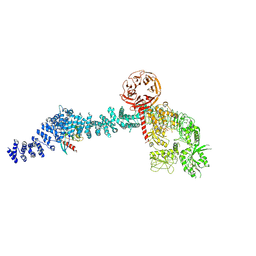 | | Cryo-EM structure of Rab12-LRRK2 complex in the LRRK2 monomer state | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2, MAGNESIUM ION, ... | | Authors: | Zhu, H, Sun, J. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-09 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | RAB12-LRRK2 complex suppresses primary ciliogenesis and regulates centrosome homeostasis in astrocytes.
Nat Commun, 15, 2024
|
|
6JZA
 
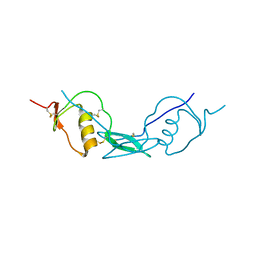 | | Structure of Fstl1 | | Descriptor: | Follistatin-related protein 1 | | Authors: | Liu, X, Ning, W. | | Deposit date: | 2019-04-30 | | Release date: | 2019-08-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional study of FK domain of Fstl1.
Protein Sci., 28, 2019
|
|
6K32
 
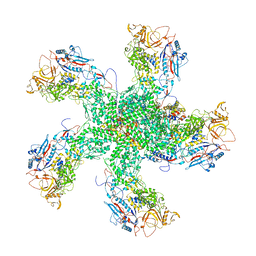 | | RdRp complex | | Descriptor: | 2'-O-methyladenosine 5'-(dihydrogen phosphate), 7-METHYLGUANOSINE, DIPHOSPHATE, ... | | Authors: | Li, X.W. | | Deposit date: | 2019-05-16 | | Release date: | 2019-11-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of RdRps Within a Transcribing dsRNA Virus Provides Insights Into the Mechanisms of RNA Synthesis.
J.Mol.Biol., 432, 2020
|
|
1BBA
 
 | |
3SRQ
 
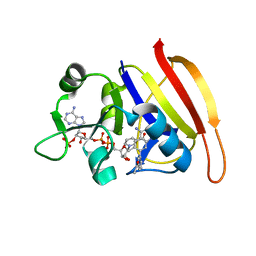 | | S. aureus Dihydrofolate Reductase complexed with novel 7-aryl-2,4-diaminoquinazolines | | Descriptor: | 1-[3-(2,4-diamino-6-methylquinazolin-7-yl)phenyl]ethanone, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M. | | Deposit date: | 2011-07-07 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-based design of new DHFR-based antibacterial agents: 7-aryl-2,4-diaminoquinazolines.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SRS
 
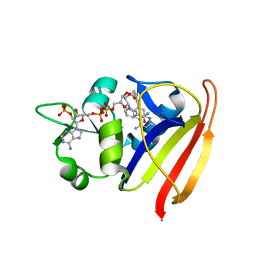 | | S. aureus Dihydrofolate Reductase complexed with novel 7-aryl-2,4-diaminoquinazolines | | Descriptor: | 7-(5-bromo-2-ethoxyphenyl)-6-methylquinazoline-2,4-diamine, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M. | | Deposit date: | 2011-07-07 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of new DHFR-based antibacterial agents: 7-aryl-2,4-diaminoquinazolines.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SQY
 
 | |
3SR5
 
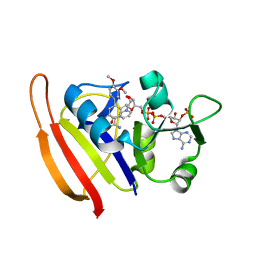 | |
3SRW
 
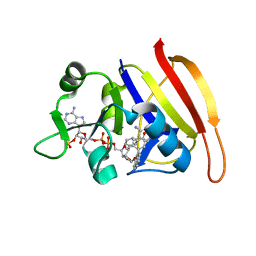 | | S. aureus Dihydrofolate Reductase complexed with novel 7-aryl-2,4-diaminoquinazolines | | Descriptor: | 7-(2-ethoxynaphthalen-1-yl)-6-methylquinazoline-2,4-diamine, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M. | | Deposit date: | 2011-07-07 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of new DHFR-based antibacterial agents: 7-aryl-2,4-diaminoquinazolines.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SRR
 
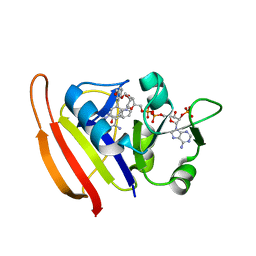 | | S. aureus Dihydrofolate Reductase complexed with novel 7-aryl-2,4-diaminoquinazolines | | Descriptor: | 3-(2,4-diamino-6-methylquinazolin-7-yl)-4-ethoxybenzaldehyde, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M. | | Deposit date: | 2011-07-07 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of new DHFR-based antibacterial agents: 7-aryl-2,4-diaminoquinazolines.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SRU
 
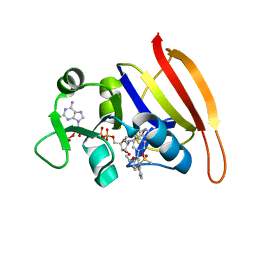 | | S. aureus Dihydrofolate Reductase complexed with novel 7-aryl-2,4-diaminoquinazolines | | Descriptor: | Dihydrofolate reductase, N-[3'-(2,4-diaminoquinazolin-7-yl)-4'-ethoxybiphenyl-3-yl]methanesulfonamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M. | | Deposit date: | 2011-07-07 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of new DHFR-based antibacterial agents: 7-aryl-2,4-diaminoquinazolines.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5ZTY
 
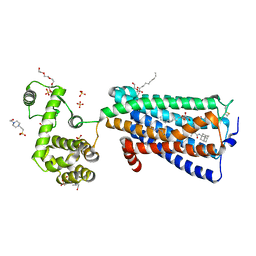 | | Crystal structure of human G protein coupled receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, X.T, Hua, T, Wu, L.J, Liu, Z.J. | | Deposit date: | 2018-05-05 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Human Cannabinoid Receptor CB2
Cell, 176, 2019
|
|
7VKL
 
 | |
4QRE
 
 | |
8AS9
 
 | | Crystal structure of the talin-KANK1 complex | | Descriptor: | B-cell lymphoma 6 protein, GLYCEROL, KN-motif NCoR1 BBD fusion,Nuclear receptor corepressor 1, ... | | Authors: | Zacharchenko, T. | | Deposit date: | 2022-08-18 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis of the talin-KANK1 interaction that coordinates the actin and microtubule cytoskeletons at focal adhesions.
Open Biology, 13, 2023
|
|
7VSJ
 
 | |
7WW3
 
 | | Crystal structure of MmIMP1-KH34 tandem domain | | Descriptor: | Insulin-like growth factor 2 mRNA-binding protein 1 | | Authors: | Li, X.J, Wu, B.X. | | Deposit date: | 2022-02-12 | | Release date: | 2023-02-22 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the RNA binding properties of mouse IGF2BP3.
Structure, 2025
|
|
