6LGL
 
 | | The atomic structure of varicella-zoster virus A-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Zheng, Q, Li, S. | | Deposit date: | 2019-12-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
8F5T
 
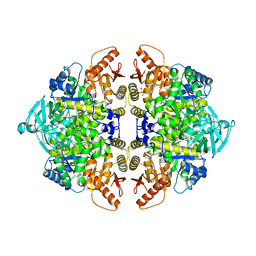 | | Rabbit muscle pyruvate kinase in complex with sodium and magnesium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Holyoak, T, Fenton, A.W. | | Deposit date: | 2022-11-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | PYK-SubstitutionOME: an integrated database containing allosteric coupling, ligand affinity and mutational, structural, pathological, bioinformatic and computational information about pyruvate kinase isozymes.
Database (Oxford), 2023, 2023
|
|
8F6M
 
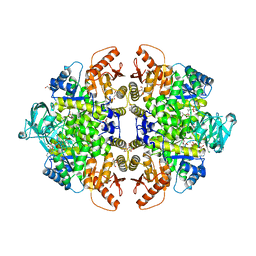 | | Complex of Rabbit muscle pyruvate kinase with ADP and the phosphonate analogue of PEP mimicking the Michaelis complex. | | Descriptor: | (E)-2-METHYL-3-PHOSPHONOACRYLATE, ADENOSINE-5'-DIPHOSPHATE, ALANINE, ... | | Authors: | Holyoak, T, Fenton, A.W. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | PYK-SubstitutionOME: an integrated database containing allosteric coupling, ligand affinity and mutational, structural, pathological, bioinformatic and computational information about pyruvate kinase isozymes.
Database (Oxford), 2023, 2023
|
|
8F5U
 
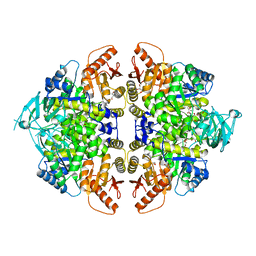 | | Rabbit muscle pyruvate kinase in complex with magnesium, potassium and pyruvate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Holyoak, T, Fenton, A.W. | | Deposit date: | 2022-11-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | PYK-SubstitutionOME: an integrated database containing allosteric coupling, ligand affinity and mutational, structural, pathological, bioinformatic and computational information about pyruvate kinase isozymes.
Database (Oxford), 2023, 2023
|
|
6MT0
 
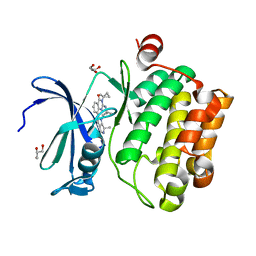 | | Crystal structure of human Pim-1 kinase in complex with a quinazolinone-pyrrolodihydropyrrolone inhibitor | | Descriptor: | 3-(1-methylcyclopropyl)-2-[(1-methylcyclopropyl)amino]-8-[(6R)-6-methyl-4-oxo-1,4,5,6-tetrahydropyrrolo[3,4-b]pyrrol-2-yl]quinazolin-4(3H)-one, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Mohr, C. | | Deposit date: | 2018-10-18 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of ( R)-8-(6-Methyl-4-oxo-1,4,5,6-tetrahydropyrrolo[3,4- b]pyrrol-2-yl)-3-(1-methylcyclopropyl)-2-((1-methylcyclopropyl)amino)quinazolin-4(3 H)-one, a Potent and Selective Pim-1/2 Kinase Inhibitor for Hematological Malignancies.
J. Med. Chem., 62, 2019
|
|
5EOL
 
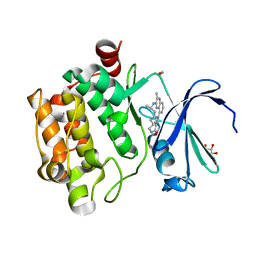 | |
6XMH
 
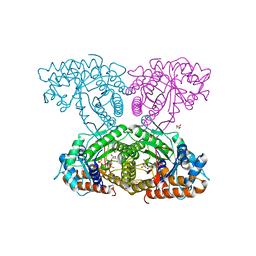 | | Human aldolase A wild type | | Descriptor: | Fructose-bisphosphate aldolase A, GLYCEROL, PHOSPHATE ION | | Authors: | Meneely, K.M, Lamb, A.L. | | Deposit date: | 2020-06-30 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substitutions at a rheostat position in human aldolase A cause a shift in the conformational population.
Protein Sci., 31, 2022
|
|
6XMM
 
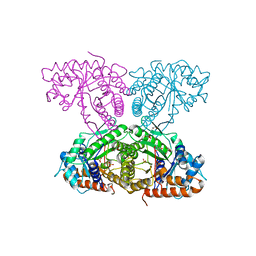 | | Human aldolase A I98S | | Descriptor: | Fructose-bisphosphate aldolase A, GLYCEROL, PHOSPHATE ION | | Authors: | Meneely, K.M, Brewer, K, Lamb, A.L. | | Deposit date: | 2020-06-30 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Substitutions at a rheostat position in human aldolase A cause a shift in the conformational population.
Protein Sci., 31, 2022
|
|
6XML
 
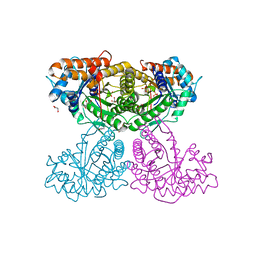 | | Human aldolase A I98C | | Descriptor: | Fructose-bisphosphate aldolase A, GLYCEROL, PHOSPHATE ION | | Authors: | Meneely, K.M, Brewer, K, Lamb, A.L. | | Deposit date: | 2020-06-30 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Substitutions at a rheostat position in human aldolase A cause a shift in the conformational population.
Protein Sci., 31, 2022
|
|
6XMO
 
 | | Human aldolase A I98F | | Descriptor: | Fructose-bisphosphate aldolase A, GLYCEROL, PHOSPHATE ION | | Authors: | Meneely, K.M, Lamb, A.L. | | Deposit date: | 2020-06-30 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substitutions at a rheostat position in human aldolase A cause a shift in the conformational population.
Protein Sci., 31, 2022
|
|
6LQA
 
 | | voltage-gated sodium channel Nav1.5 with quinidine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Quinidine, Sodium channel protein type 5 subunit alpha | | Authors: | Yan, N, Li, Z, Pan, X, Huang, G. | | Deposit date: | 2020-01-13 | | Release date: | 2021-03-24 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Basis for Pore Blockade of the Human Cardiac Sodium Channel Na v 1.5 by the Antiarrhythmic Drug Quinidine*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7KQP
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ADP-ribose (P43 crystal form) | | Descriptor: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-17 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KQW
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, methylated) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-17 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KQO
 
 | |
7KR1
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 310 K) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7KR0
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, 100 K) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.77 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5IPJ
 
 | | Crystal structure of human Pim-1 kinase in complex with a quinazolinone-pyrrolopyrrolone inhibitor. | | Descriptor: | 2-(tert-butylamino)-3-methyl-8-[(6R)-6-methyl-4-oxo-1,4,5,6-tetrahydropyrrolo[3,4-b]pyrrol-2-yl]quinazolin-4(3H)-one, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Mohr, C. | | Deposit date: | 2016-03-09 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Optimization of Quinazolinone-pyrrolopyrrolones as Potent and Orally Bioavailable Pan-Pim Kinase Inhibitors.
J.Med.Chem., 59, 2016
|
|
3QJZ
 
 | | Crystal structure of PI3K-gamma in complex with benzothiazole 1 | | Descriptor: | N-{6-[2-(methylsulfanyl)pyrimidin-4-yl]-1,3-benzothiazol-2-yl}acetamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2011-01-31 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery and Optimization of a Series of Benzothiazole Phosphoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QK0
 
 | | Crystal structure of PI3K-gamma in complex with benzothiazole 82 | | Descriptor: | N-[6-(6-chloro-5-{[(4-fluorophenyl)sulfonyl]amino}pyridin-3-yl)-1,3-benzothiazol-2-yl]acetamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2011-01-31 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery and Optimization of a Series of Benzothiazole Phosphoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors.
J.Med.Chem., 54, 2011
|
|
8S9M
 
 | | DNA cytosine-N4 methyltransferase (residues 79-324) from the Bdelloid rotifer Adineta vaga | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
8S9N
 
 | | DNA cytosine-N4 methyltransferase (residues 61-324) from the Bdelloid rotifer Adineta vaga - C2 crystal form | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
8S9O
 
 | | DNA cytosine-N4 methyltransferase (residues 61-324) from the Bdelloid rotifer Adineta vaga - P1 crystal form | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
6X00
 
 | | Structure of DbNA(11) peptides bound to H-2Db MHC-I | | Descriptor: | Beta-2-microglobulin, Epitope from Neuraminidase Protein (NA-181-191), H-2 class I histocompatibility antigen, ... | | Authors: | Farenc, C, Rossjohn, J. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Overlapping Peptides Elicit Distinct CD8 + T Cell Responses following Influenza A Virus Infection.
J Immunol., 205, 2020
|
|
6WZY
 
 | | Structure of DbNA(10) peptides bound to H-2Db MHC-I | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Farenc, C, Rossjohn, J. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Overlapping Peptides Elicit Distinct CD8 + T Cell Responses following Influenza A Virus Infection.
J Immunol., 205, 2020
|
|
7WP6
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 36H6 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-01 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
