6YDF
 
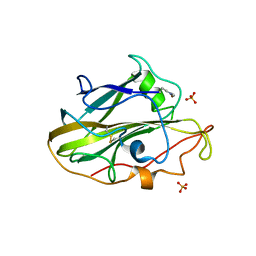 | | X-ray structure of LPMO. | | Descriptor: | COPPER (II) ION, LPMO lytic polysaccharide monooxygenase, SULFATE ION | | Authors: | Tandrup, T, Tryfona, T, Frandsen, K.E.H, Johansen, K.S, Dupree, P, Lo Leggio, L. | | Deposit date: | 2020-03-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Oligosaccharide Binding and Thermostability of Two Related AA9 Lytic Polysaccharide Monooxygenases.
Biochemistry, 59, 2020
|
|
8PNJ
 
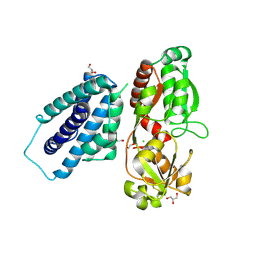 | | Chorismate mutase | | Descriptor: | 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Khatanbaatar, T, Cordara, G, Krengel, U. | | Deposit date: | 2023-06-30 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.355 Å) | | Cite: | Structural analysis of chorismate mutase and cyclohexadienyl dehydratase from Pseudomonas aeruginosa
To Be Published
|
|
8PNH
 
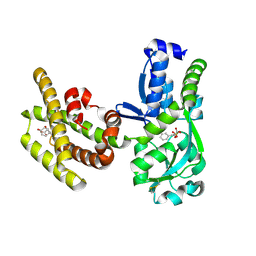 | | Chorismate mutase | | Descriptor: | 3-PHENYLPYRUVIC ACID, 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, Bifunctional cyclohexadienyl dehydratase/chorismate mutase from Janthinobacterium sp. HH01, ... | | Authors: | Khatanbaatar, T, Cordara, G, Krengel, U. | | Deposit date: | 2023-06-30 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chorismate mutase
To Be Published
|
|
8PNI
 
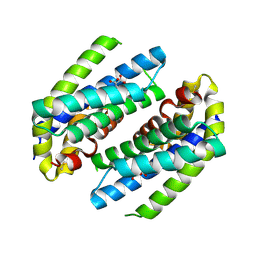 | | Chorismate mutase | | Descriptor: | CITRIC ACID, Monofunctional chorismate mutase | | Authors: | Khatanbaatar, T, Cordara, G, Krengel, U. | | Deposit date: | 2023-06-30 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural analysis of chorismate mutase and cyclohexadienyl dehydratase from Pseudomonas aeruginosa
To Be Published
|
|
7PXV
 
 | | LsAA9_A chemically reduced with ascorbic acid (high X-ray dose) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PZ7
 
 | | Structure of an LPMO at 1.13x10^6 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACRYLIC ACID, ... | | Authors: | Tandrup, T, Muderspach, S.J, Ipsen, J.O, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PXM
 
 | | X-ray structure of LPMO at 1.45x10^6 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, COPPER (II) ION | | Authors: | Tandrup, T, Lo Leggio, L. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PYW
 
 | | Structure of LPMO (expressed in E.coli) with cellotriose at 5.62x10^4 Gy | | Descriptor: | ACETATE ION, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
6YDD
 
 | | X-ray structure of LPMO. | | Descriptor: | COPPER (II) ION, LPMO lytic polysaccharide monooxygenase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Tandrup, T, Tryfona, T, Frandsen, K.E.H, Johansen, K.S, Dupree, P, Lo Leggio, L. | | Deposit date: | 2020-03-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Oligosaccharide Binding and Thermostability of Two Related AA9 Lytic Polysaccharide Monooxygenases.
Biochemistry, 59, 2020
|
|
5H7K
 
 | | Crystal structure of Elongation factor 2 GDP-form | | Descriptor: | Elongation factor 2, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Tanzawa, T, Kato, K, Uchiumi, T, Yao, M. | | Deposit date: | 2016-11-18 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | The C-terminal helix of ribosomal P stalk recognizes a hydrophobic groove of elongation factor 2 in a novel fashion
Nucleic Acids Res., 46, 2018
|
|
8IK2
 
 | | RhlA exhibits dual thioesterase and acyltransferase activities during rhamnolipid biosynthesis | | Descriptor: | (3~{S})-3-oxidanyldecanoic acid, 3-(3-hydroxydecanoyloxy)decanoate synthase | | Authors: | Tang, T, Fu, L.H, Xie, W.H, Luo, Y.Z, Zhang, Y.T, Si, T. | | Deposit date: | 2023-02-28 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | RhlA Exhibits Dual Thioesterase and Acyltransferase Activities during Rhamnolipid Biosynthesis
Acs Catalysis, 13, 2023
|
|
6YGT
 
 | |
3X3U
 
 | |
1V4R
 
 | | Solution structure of Streptmycal repressor TraR | | Descriptor: | Transcriptional Repressor | | Authors: | Tanaka, T, Komatsu, C, Kobayashi, K, Sugai, M, Kataoka, M, Kohno, T. | | Deposit date: | 2003-11-17 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Streptmycal repressor TraR
TO BE PUBLISHED
|
|
7CLJ
 
 | | Crystal structure of Thermoplasmatales archaeon heliorhodopsin E108D mutant | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, RETINAL, SULFATE ION, ... | | Authors: | Tanaka, T, Shihoya, W, Yamashita, K, Nureki, O. | | Deposit date: | 2020-07-21 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for unique color tuning mechanism in heliorhodopsin.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7Y4P
 
 | | Human Plexin A1, extracellular domains 1-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A1 | | Authors: | Tanaka, T, Neyazaki, M, Nogi, T. | | Deposit date: | 2022-06-15 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Hybrid in vitro/in silico analysis of low-affinity protein-protein interactions that regulate signal transduction by Sema6D.
Protein Sci., 31, 2022
|
|
7Y4O
 
 | | Rat Semaphorin 6D extracellular region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Semaphorin 6D | | Authors: | Tanaka, T, Neyazaki, M, Nogi, T. | | Deposit date: | 2022-06-15 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Hybrid in vitro/in silico analysis of low-affinity protein-protein interactions that regulate signal transduction by Sema6D.
Protein Sci., 31, 2022
|
|
7Y4Q
 
 | | Semaphorin 6D in complex with Plexin A1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A1, ... | | Authors: | Tanaka, T, Neyazaki, M, Nogi, T. | | Deposit date: | 2022-06-16 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.7 Å) | | Cite: | Hybrid in vitro/in silico analysis of low-affinity protein-protein interactions that regulate signal transduction by Sema6D.
Protein Sci., 31, 2022
|
|
3G3N
 
 | | PDE7A catalytic domain in complex with 3-(2,6-difluorophenyl)-2-(methylthio)quinazolin-4(3H)-one | | Descriptor: | 3-(2,6-difluorophenyl)-2-(methylthio)quinazolin-4(3H)-one, High affinity cAMP-specific 3',5'-cyclic phosphodiesterase 7A, MAGNESIUM ION, ... | | Authors: | Castano, T, Wang, H. | | Deposit date: | 2009-02-02 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis, structural analysis, and biological evaluation of thioxoquinazoline derivatives as phosphodiesterase 7 inhibitors
Chemmedchem, 4, 2009
|
|
1IKU
 
 | | myristoylated recoverin in the calcium-free state, NMR, 22 structures | | Descriptor: | MYRISTIC ACID, RECOVERIN | | Authors: | Tanaka, T, Ames, J.B, Harvey, T.S, Stryer, L, Ikura, M. | | Deposit date: | 1996-01-18 | | Release date: | 1996-07-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Sequestration of the membrane-targeting myristoyl group of recoverin in the calcium-free state.
Nature, 376, 1995
|
|
7KZ6
 
 | | Crystal structure of KabA from Bacillus cereus UW85 with bound cofactor PMP | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme | | Authors: | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
7KZ3
 
 | | Crystal structure of KabA from Bacillus cereus UW85 in complex with the internal aldimine | | Descriptor: | 1,2-ETHANEDIOL, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, SODIUM ION | | Authors: | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2020-12-09 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
7KZ5
 
 | | Crystal structure of KabA from Bacillus cereus UW85 in complex with the plp external aldimine adduct with kanosamine-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 3-deoxy-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-O-phosphono-alpha-D-gluco pyranose, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, ... | | Authors: | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
7KZD
 
 | | Crystal structure of KabA from Bacillus cereus UW85 in complex with the reduced internal aldimine and with bound Glutarate | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, 1,2-ETHANEDIOL, Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, ... | | Authors: | Prasertanan, T, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Snapshots along the catalytic path of KabA, a PLP-dependent aminotransferase required for kanosamine biosynthesis in Bacillus cereus UW85.
J.Struct.Biol., 213, 2021
|
|
2DUU
 
 | | Crystal Structure of apo-form of NADP-Dependent Glyceraldehyde-3-Phosphate Dehydrogenase from Synechococcus Sp. | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase, SULFATE ION | | Authors: | Kitatani, T, Nakamura, Y, Wada, K, Kinoshita, T, Tamoi, M, Shigeoka, S, Tada, T. | | Deposit date: | 2006-07-27 | | Release date: | 2006-09-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of apo-glyceraldehyde-3-phosphate dehydrogenase from Synechococcus PCC7942
Acta Crystallogr.,Sect.F, 62, 2006
|
|
