7NRE
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with darobactin (crystal form 1) | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Darobactin, MAGNESIUM ION, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
7NRI
 
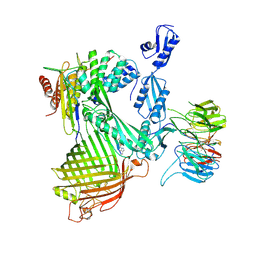 | | Structure of the darobactin-bound E. coli BAM complex (BamABCDE) | | Descriptor: | 3-PYRIDIN-4-YL-2,4-DIHYDRO-INDENO[1,2-.C.]PYRAZOLE, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Jakob, R.P, Kaur, H, Marzinek, J.K, Green, R, Imai, Y, Bolla, J, Robinson, C, Bond, P.J, Lewis, K, Maier, T, Hiller, S. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-21 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase.
Nature, 593, 2021
|
|
7NGQ
 
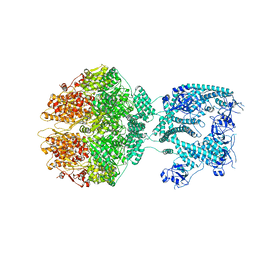 | | Human mitochondrial Lon protease homolog, D2-state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Mohammed, I, Abrahams, J.P, Schmitz, K.A, Maier, T, Schenck, N. | | Deposit date: | 2021-02-09 | | Release date: | 2021-04-28 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
7NGP
 
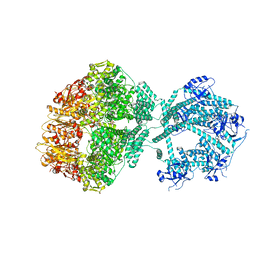 | | D1-state of wild type human mitochondrial LONP1 protease | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T, Abrahams, J.P. | | Deposit date: | 2021-02-09 | | Release date: | 2021-04-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
7NGF
 
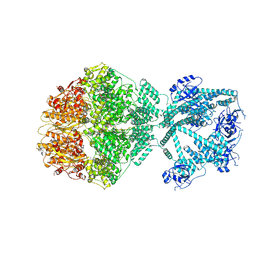 | | P2c-state of wild type human mitochondrial LONP1 protease with bound endogenous substrate protein and in presence of ATP/ADP mix | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Lon protease homolog, ... | | Authors: | Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T, Abrahams, J.P. | | Deposit date: | 2021-02-09 | | Release date: | 2021-04-28 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
7NGL
 
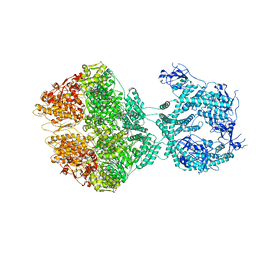 | | R-state of wild type human mitochondrial LONP1 protease bound to endogenous ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Mohammed, I, Schmitz, K.A, Schenck, N, Maier, T, Abrahams, J.P. | | Deposit date: | 2021-02-09 | | Release date: | 2021-04-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Catalytic cycling of human mitochondrial Lon protease.
Structure, 30, 2022
|
|
7AYN
 
 | | Crystal structure of the lectin domain of the FimH variant Arg98Ala, in complex with Methyl 3-chloro-4-D-mannopyranosyloxy-3-biphenylcarboxylate | | Descriptor: | 1,2-ETHANEDIOL, Type 1 fimbrin D-mannose specific adhesin, methyl 3-[3-chloranyl-4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-phenyl]benzoate | | Authors: | Jakob, R.P, Tomasic, T, Rabbani, S, Reisner, A, Jakopin, Z, Maier, T, Ernst, B, Anderluh, M. | | Deposit date: | 2020-11-12 | | Release date: | 2020-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Does targeting Arg98 of FimH lead to high affinity antagonists?
Eur.J.Med.Chem., 211, 2020
|
|
6QGY
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody B12 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoB12, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
1W26
 
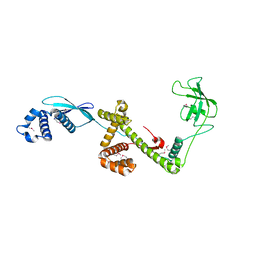 | | Trigger Factor in Complex with the Ribosome forms a Molecular Cradle for Nascent Proteins | | Descriptor: | TRIGGER FACTOR | | Authors: | Ferbitz, L, Maier, T, Patzelt, H, Bukau, B, Deuerling, E, Ban, N. | | Deposit date: | 2004-06-28 | | Release date: | 2004-09-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Trigger Factor in Complex with the Ribosome Forms a Molecular Cradle for Nascent Proteins
Nature, 431, 2004
|
|
1W2B
 
 | | Trigger Factor ribosome binding domain in complex with 50S | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L13P, ... | | Authors: | Ferbitz, L, Maier, T, Patzelt, H, Bukau, B, Deuerling, E, Ban, N. | | Deposit date: | 2004-07-01 | | Release date: | 2004-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Trigger Factor in Complex with the Ribosome Forms a Molecular Cradle for Nascent Proteins
Nature, 431, 2004
|
|
8CG5
 
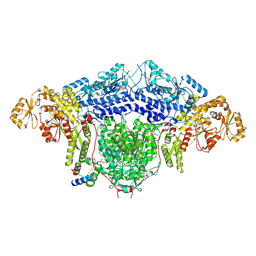 | |
8CG4
 
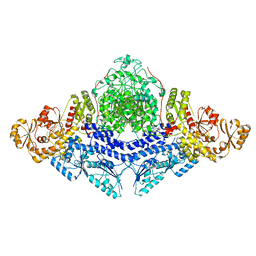 | |
8BP6
 
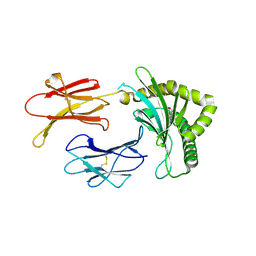 | | Structure of MHC-class I related molecule MR1 with bound M3Ade. | | Descriptor: | (1R,5S)-8-(9H-purin-6-yl)-2-oxa-8-azabicyclo[3.3.1]nona-3,6-diene-4,6-dicarbaldehyde, Beta-2-microglobulin,Major histocompatibility complex class I-related gene protein | | Authors: | Berloffa, G, Jakob, R.P, Maier, T. | | Deposit date: | 2022-11-16 | | Release date: | 2023-11-29 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The carbonyl nucleobase adduct M 3 Ade is a potent antigen for adaptive polyclonal MR1-restricted T cells.
Immunity, 2024
|
|
8BY8
 
 | |
8CG6
 
 | |
7ZMC
 
 | |
7ZMA
 
 | |
7ZM9
 
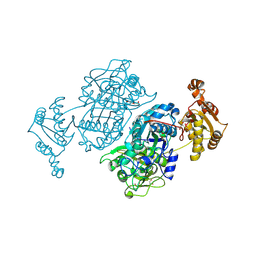 | | Ketosynthase domain 3 of Brevibacillus Brevis orphan BGC11 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Putative polyketide synthase | | Authors: | Tittes, Y.U, Herbst, D.A, Jakob, R.P, Maier, T. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The structure of a polyketide synthase bimodule core.
Sci Adv, 8, 2022
|
|
7ZMF
 
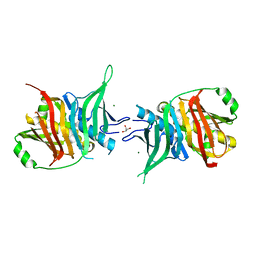 | | Dehydratase domain of module 3 from Brevibacillus Brevis orphan BGC11 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative polyketide synthase | | Authors: | Tittes, Y.U, Herbst, D.A, Jakob, R.P, Maier, T. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The structure of a polyketide synthase bimodule core.
Sci Adv, 8, 2022
|
|
7ZMD
 
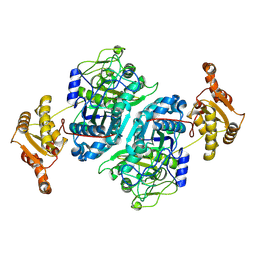 | |
7ZSK
 
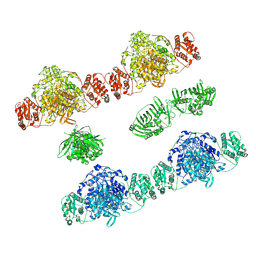 | |
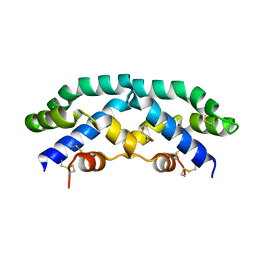
2Z9A
 
 | |
2VRH
 
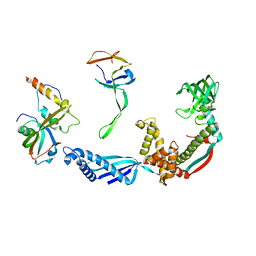 | | Structure of the E. coli trigger factor bound to a translating ribosome | | Descriptor: | 50S RIBOSOMAL PROTEIN L23, 50S RIBOSOMAL PROTEIN L24, 50S RIBOSOMAL PROTEIN L29, ... | | Authors: | Merz, F, Boehringer, D, Schaffitzel, C, Preissler, S, Hoffmann, A, Maier, T, Rutkowska, A, Lozza, J, Ban, N, Bukau, B, Deuerling, E. | | Deposit date: | 2008-04-07 | | Release date: | 2008-06-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | Molecular Mechanism and Structure of Trigger Factor Bound to the Translating Ribosome.
Embo J., 27, 2008
|
|
2WCD
 
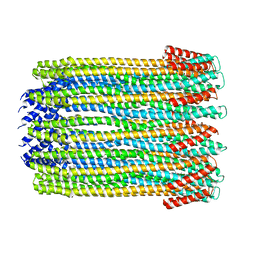 | | Crystal structure of the assembled cytolysin A pore | | Descriptor: | ETHYL MERCURY ION, HEMOLYSIN E, CHROMOSOMAL | | Authors: | Mueller, M, Grauschopf, U, Maier, T, Glockshuber, R, Ban, N. | | Deposit date: | 2009-03-11 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | The Structure of a Cytolytic Alpha-Helical Toxin Pore Reveals its Assembly Mechanism
Nature, 459, 2009
|
|
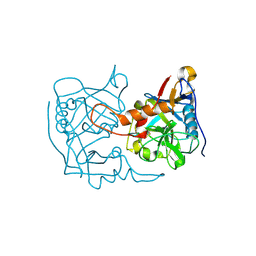
5EX1
 
 | |
