5WQQ
 
 | | High resolution structure of high-potential iron-sulfur protein in the oxidized state | | Descriptor: | GLYCEROL, High-potential iron-sulfur protein, IRON/SULFUR CLUSTER, ... | | Authors: | Ohno, H, Takeda, K, Niwa, S, Tsujinaka, T, Hanazono, Y, Hirano, Y, Miki, K. | | Deposit date: | 2016-11-28 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Crystallographic characterization of the high-potential iron-sulfur protein in the oxidized state at 0.8 angstrom resolution
PLoS ONE, 12, 2017
|
|
4IN0
 
 | |
3RN5
 
 | | Structural basis of cytosolic DNA recognition by innate immune receptors | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*CP*AP*AP*AP*GP*AP*GP*AP*GP*AP*AP*AP*GP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*CP*TP*TP*TP*CP*TP*CP*TP*CP*TP*TP*TP*GP*AP*TP*G)-3'), ... | | Authors: | Jin, T.C, Xiao, T. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
3RNU
 
 | | Structural Basis of Cytosolic DNA Sensing by Innate Immune Receptors | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*CP*AP*TP*CP*AP*AP*AP*GP*AP*GP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*CP*TP*TP*TP*GP*AP*TP*GP*GP*CP*C)-3'), ... | | Authors: | Jin, T.C, Xiao, T. | | Deposit date: | 2011-04-22 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
3RN2
 
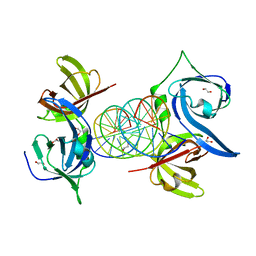 | | Structural Basis of Cytosolic DNA Recognition by Innate Immune Receptors | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*CP*AP*AP*AP*GP*AP*TP*CP*TP*TP*TP*GP*AP*TP*GP*G)-3'), Interferon-inducible protein AIM2 | | Authors: | Jin, T.C, Xiao, T. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
3RLN
 
 | |
3RLO
 
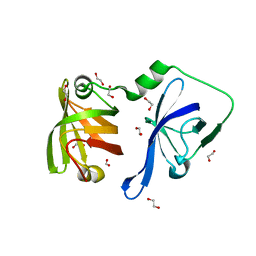 | | Structural Basis of Cytosolic DNA Recognition by Innate Receptors | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Gamma-interferon-inducible protein 16 | | Authors: | Jin, T.C, Xiao, T. | | Deposit date: | 2011-04-19 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
2RE9
 
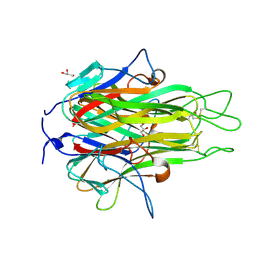 | | Crystal structure of TL1A at 2.1 A | | Descriptor: | GLYCEROL, MAGNESIUM ION, TNF superfamily ligand TL1A | | Authors: | Jin, T.C, Guo, F, Kim, S, Howard, A.J, Zhang, Y.Z. | | Deposit date: | 2007-09-25 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of TNF ligand family member TL1A at 2.1 A.
Biochem.Biophys.Res.Commun., 364, 2007
|
|
3FZ3
 
 | | Crystal Structure of almond Pru1 protein | | Descriptor: | CALCIUM ION, Prunin, SODIUM ION | | Authors: | Jin, T.C, Zhang, Y.Z. | | Deposit date: | 2009-01-23 | | Release date: | 2009-11-10 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of prunin-1, a major component of the almond (Prunus dulcis) allergen amandin.
J.Agric.Food Chem., 57, 2009
|
|
5H6I
 
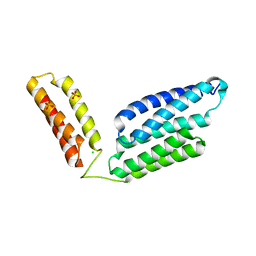 | | Crystal Structure of GBS CAMP Factor | | Descriptor: | CHLORIDE ION, Protein B, SULFATE ION | | Authors: | Jin, T.C, Brefo-Mensah, E.K. | | Deposit date: | 2016-11-13 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of theStreptococcus agalactiaeCAMP factor provides insights into its membrane-permeabilizing activity.
J.Biol.Chem., 293, 2018
|
|
4LEJ
 
 | | Crystal Structure of the Korean pine (Pinus koraiensis) vicilin | | Descriptor: | COPPER (II) ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Jin, T.C, Wang, Y, Zhang, Y.Z. | | Deposit date: | 2013-06-25 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Crystal Structure of Korean Pine ( Pinus koraiensis ) 7S Seed Storage Protein with Copper Ligands.
J.Agric.Food Chem., 62, 2014
|
|
2O0O
 
 | | Crystal structure of TL1A | | Descriptor: | MAGNESIUM ION, TNF superfamily ligand TL1A | | Authors: | Jin, T.C, Kim, S, Guo, F, Howard, A.J, Zhang, Y.Z. | | Deposit date: | 2006-11-27 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray crystal structure of TNF ligand family member TL1A at 2.1A.
Biochem.Biophys.Res.Commun., 364, 2007
|
|
3VD8
 
 | | Crystal structure of human AIM2 PYD domain with MBP fusion | | Descriptor: | 1,2-ETHANEDIOL, Maltose-binding periplasmic protein, Interferon-inducible protein AIM2, ... | | Authors: | Jin, T.C, Perry, A, Smith, P, Xiao, T.S. | | Deposit date: | 2012-01-04 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0685 Å) | | Cite: | Structure of the Absent in Melanoma 2 (AIM2) Pyrin Domain Provides Insights into the Mechanisms of AIM2 Autoinhibition and Inflammasome Assembly.
J.Biol.Chem., 288, 2013
|
|
2NML
 
 | | Crystal structure of HEF2/ERH at 1.55 A resolution | | Descriptor: | Enhancer of rudimentary homolog | | Authors: | Jin, T.C, Guo, F, Serebriiskii, I.G, Howard, A.J, Zhang, Y.Z. | | Deposit date: | 2006-10-21 | | Release date: | 2006-10-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A 1.55 A resolution X-ray crystal structure of HEF2/ERH and insights into its transcriptional and cell-cycle interaction networks.
Proteins, 68, 2007
|
|
8I6K
 
 | | Structure of hMNDA HIN with dsDNA | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Li, Y.L, Jin, T.C. | | Deposit date: | 2023-01-28 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural mechanism of dsDNA recognition by the hMNDA HIN domain: New insights into the DNA-binding model of a PYHIN protein.
Int.J.Biol.Macromol., 245, 2023
|
|
6IWK
 
 | | The Structure of Maltooligosaccharide-forming Amylase from Pseudomonas saccharophila STB07 | | Descriptor: | CALCIUM ION, GLYCEROL, Glucan 1,4-alpha-maltotetraohydrolase | | Authors: | Li, Z.F, Ban, X.F, Zhang, Z.Q, Li, C.M, Gu, Z.B, Jin, T.C, Li, Y.L, Shang, Y.H. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structure of maltotetraose-forming amylase from Pseudomonas saccharophila STB07 provides insights into its product specificity.
Int.J.Biol.Macromol., 154, 2020
|
|
6UUO
 
 | | Crystal structure of BRAF kinase domain bound to the PROTAC P4B | | Descriptor: | N-(3-{5-[(1-acetylpiperidin-4-yl)(methyl)amino]-3-(pyrimidin-5-yl)-1H-pyrrolo[3,2-b]pyridin-1-yl}-2,4-difluorophenyl)propane-1-sulfonamide, Serine/threonine-protein kinase B-raf | | Authors: | Maisonneuve, P, Posternak, G, Kurinov, I, Sicheri, F. | | Deposit date: | 2019-10-30 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.288 Å) | | Cite: | Functional characterization of a PROTAC directed against BRAF mutant V600E.
Nat.Chem.Biol., 16, 2020
|
|
5YGP
 
 | | Human TNFRSF25 death domain mutant-D412E | | Descriptor: | SULFATE ION, TNFRSF25 death domain, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yin, X, Jin, T.C. | | Deposit date: | 2017-09-25 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure and activation mechanism of DR3 death domain.
Febs J., 286, 2019
|
|
7V4Q
 
 | |
7VOA
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with aRBD5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpaca nanobody | | Authors: | Ma, H, Zeng, W.H, Jin, T.C. | | Deposit date: | 2021-10-13 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hetero-bivalent nanobodies provide broad-spectrum protection against SARS-CoV-2 variants of concern including Omicron.
Cell Res., 32, 2022
|
|
7V4R
 
 | | The crystal structure of KFDV NS3H bound with Pi | | Descriptor: | NICKEL (II) ION, PHOSPHATE ION, Serine protease NS3 | | Authors: | Zhang, C.Y, Jin, T.C. | | Deposit date: | 2021-08-14 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kyasanur Forest disease virus NS3 helicase: Insights into structure, activity, and inhibitors.
Int.J.Biol.Macromol., 2023
|
|
7ESV
 
 | |
7ESQ
 
 | |
7ESP
 
 | |
7ESO
 
 | |
